ABSTRACT
Staphylococcus aureus is commonly involved in food poisoning due to production of toxins responsible for causing animal and human diseases. In this study, 60 strains of presumptive S. aureus isolates from raw milk and cheese were biochemically identified in four dairies: 54 (90%) from refrigerated raw milk (RRM) with counts exceeding 106 CFU/mL, and six (10%) from cheese with similar concentrations of CFU/mL. Out of the 60 strains of presumptive S. aureus, 46 (76.7%) amplified the femA gene and then they were investigated regarding the presence of the Toxic Shock Syndrome Toxin-1 (TSST-1) gene and the classical enterotoxin genes (SEs) types A, B, C, D and E: 31 (67.4%) carried one or more encoding toxin genes, and 13 different genotypes were identified. Twenty-one strains (61.8%) carried one gene; three (8.8%), two genes; seven (20.6%), three genes; two (5.8%), four genes; and one (3%), five genes. The sec gene was the most frequent one, followed by seb and tst. The sed gene was expressed by 10 strains (29.4%), sea by five (14.7%) and see by three (8.8%). The S. aureus isolates showed genetic potential for producing toxins of importance for public health that presented a danger of food poisoning.
Key words: Staphylococcus aureus, milk, cheese, staphylococcal toxins.
Milk and cheese are widely consumed and appreciated foods around the world. Mozzarella cheese is widely used in Brazilian cuisine in hot dishes and sandwiches and coalho cheese is a popular dairy product consumed in the Northeast region of Brazil (Andreatta et al., 2009, Silva et al., 2012). Because cow’s milk contains lipids, proteins, amino acids, vitamins and minerals (Haug et al., 2007) it is considered to be an excellent culture medium for development of a variety of microorganisms. Among the main microorganisms that contaminate milk and dairy products, staphylococci stand out. The presence of this pathogen in the mammary glands of cows, especially as the etiologic agent of bovine mastitis, makes milk and dairy products great vehicles for its dispersion in Brazil (Martin et al., 2016). Staphylococcal poisoning is the most frequent cause of food-borne disease (FBD) outbreaks in many countries (Kadariya et al., 2014). It occurs after ingestion of food containing preformed enterotoxins, and raw milk, pasteurized milk and cheeses can be highlighted as the dairy products most incriminated (Le Loir et al., 2003; Oliver et al., 2005).
CDC estimates that each year roughly 48 million people gets sick from a food-borne disease (FBD) (Scharff et al., 2016). Food handlers represent an unquestionable link in the epidemiological food poisoning chain. They also play the role of a hygienic-sanitary indicator in the food industry, since they represent the main source of propagation (Kadariya et al., 2014).
Several studies have reported highest prevalence of CPS in dairy products in Brazil (Rall et al., 2008; Moraes et al., 2009; Guimaraes et al., 2013; Nunes and Caldas, 2017).
Staphylococci can produce several toxins. Among them are the classical staphylococcal enterotoxins (SEs) (SEA, SEB, SEC, SED and SEE), which are responsible for most food poisoning cases presenting clinical conditions of vomiting, diarrhea, nausea and generalized debility. They can also produce Toxic Shock Syndrome Toxin-1 (TSST-1), which is responsible for multisystemic disorders and can lead to death due to lethal shock if not properly treated (Ortega et al., 2010).
The first aim of this study was to quantify coagulase-positive Stafilococci (CPS) in the production lines of dairies producing cheese and identify S. aureus isolates by biochemical and molecular methods. The second objective was to analyze the frequencies of the genes that encode for production of the classical SEs (sea, seb, sec, sed and see) and for TSST-1 (tst) in isolated strains of S. aureus isolates.
Sampling procedure
One hundred and twenty samples were collected from four dairies, called A, B, C and D, in the state of Maranhão, Brazil. All factories were inspected by the state or federal inspection service, and produced cheese of mozzarella or coalho type from pasteurized milk.
Each dairy was visited five times, so the samples were taken from five different production batches. During each visit, two samples of refrigerated raw milk (RRM), two samples of pasteurized milk (PM) and two samples of coalho cheese (dairy A) or mozzarella cheese (dairies B, C and D) were taken. In total, 40 samples were of RRM, 40 of PM, and 40 of cheese (30 of mozzarella and 10 of coalho). The samples of refrigerated raw milk and pasteurized milk were taken from the reception tank and from the pasteurizer outlet, respectively, and kept in sterile 250 mL flasks; the cheese samples were taken after they were wrapped, in portions of 250 g. All samples were taken to the Food and Water Microbiology Laboratory in cool boxes and kept for 2 to 12 h at <4°C until microbiological analysis.
Quantification of coagulase-positive staphylococcus and biochemical identification of S. aureus isolates
To quantify coagulase-positive Staphylococcus (CPS) and isolate S. aureus, serial dilutions (10-1, 10-2, 10-3) of each sample were inoculated in dishes with Baird-Parker agar (Himedia), supplemented with egg yolk and potassium tellurite (Himedia), and were incubated at 37ºC for 48 h. After this period, typical colonies were counted and tested for Gram staining, catalase and free coagulase; the positive samples were identified as presumptive coagulase-positive Staphylococci. To biochemically identify S. aureus, all the CPS colonies were tested for Voges-Proskauer (VP) reaction and maltose and trehalose fermentation. The colonies with positive results were identified as presumptive S. aureus (Garcia, 2010).
Molecular confirmation of presumptive S. aureus isolates and toxigenic genes investigation
The isolates of S. aureus were grown in BHI broth, at 37°C/24 h. After the incubation period, 1 mL of each growth was transferred to 1.5 mL microcentrifuge tubes (Axygen) and DNA extraction was performed using the Wizard Genomic DNA Purification (Promega) commercial kit according to the manufacturer’s instructions for Gram-positive bacterial genomic DNA extraction. The extracted DNA was stored at -20ºC until its use.
Amplification of the 132 bp fragment of the femA gene (Mehrotra et al., 2000) was performed by PCR to confirm the biochemical identification of S. aureus isolates.
The reaction was carried out in a final volume of 25 µL, with 2.5 µL of 10x reaction buffer (100 mM Tris-HCl at pH 8.3 and 500 mM KCl), 2 mM of MgCl2, 200 µM of each dNTP, 20 pmol of each of the oligonucleotide primers femA-1 and femA-2 (Table 1), 2.5 U of Taq DNA polymerase (Invitrogen, Brazil) and 5 µL of the template DNA at a concentration of approximately 200 hg/μL.
The DNA amplification was performed in a Biocycler thermal cycler, under the following conditions: initial denaturation at 94ºC for five minutes, 35 amplification cycles (denaturation at 94ºC for two minutes, annealing at 57ºC for two minutes and extension at 72ºC for one minute) and a final extension at 72ºC for seven minutes.
In order to investigate the SE genes (sea, seb, sec, sed and see) a multiplex PCR was performed in a final volume of 50 µL was performed according Becker et al. (1998). In short, we used 5 µL of 10x reaction buffer (100 mM Tris-HCl at pH 8.3 and 500 mM KCl), 3 mM of MgCl2, 160 µM of each dNTP, 20 pmol of each of the SEA, SEB, SEC, SED and SEE primers (Table 1), 1.2 U of Taq DNA polymerase (Invitrogen, Brazil) and 5 µL of the template DNA at a concentration of approximately 200 hg/μL. The DNA of S. aureus ATCC 13565 (SEA), ATCC 14458 (SEB), ATCC 19095 (SEC), ATCC 23235 (SED) and ATCC 27664 (SEE), provided by Fundação Oswaldo Cruz (FIOCRUZ), were used as positive control.
To investigate the tst gene, PCR reaction was performed in a final volume of 25 µL according Mehrotra et al. (2000). In short 2.5 µL of 10x reaction buffer 10x (100 mM Tris-HCl was used at pH 8.3 and 500 mM KCl), 2 mM of MgCl2, 200 µM of each dNTP, 20 pmol of each oligonucleotide primer (TSST-1 and TSST-2), 2.5 U of Taq DNA polymerase (Invitrogen, Brazil) and 5 µL of the template DNA at a concentration of approximately 200 hg/μL.
From all the reactions, ten microliters of the amplified product were loaded onto 1% agarose gel with ethidium bromide (10 mg/mL) and underwent electrophoresis in TBE buffer (0.09 M Tris-HCl, 0.09 M boric acid and 2 mM EDTA, at pH 8.0), at 150 V for two hours. The amplified DNA was observed under ultraviolet light and the images were digitalized. A 100 bp ladder (In vitrogen, Brazil) was used as standard molecular weight.
Quantification of coagulase-positive Staphylococcus (CPS) and identification of S. aureus
CPS counts and identification of S. aureus are reported in Table 2. For CPS quantification, results are expressed as mean of two samples for each food. High CPS concentrations were observed in all 40 samples (100%) of RRM, with counts ranging from 1.7 × 104 to >106 CFU/mL. Twenty-six samples (65%) shown S. aureus contamination. From this total, 54 strains of the pathogen were biochemically identified.
In pasteurized milk S. aureus was not isolated but 8 (20%) out of the 40 samples analyzed presented CPS contamination with counts as high as 1.6 × 104 CFU/mL. All 40 (100%) of the mozzarella and coalho cheese samples also presented CPS contamination, with counts between 2.7 × 103 and >106 CFU/g. Six isolates of S. aureus were identified by biochemical tests, five from mozzarella cheese and one from coalho cheese.
Sixty presumptive S. aureus isolates were investigated by PCR in order to confirm the identification and search for toxins genes. The effectiveness of the molecular protocols is shown in Figure 1a and b. The individual amplification of each gene investigated was observed (femA, sea, seb, sec, sed, see and tst), as well as the specific and simultaneous amplification of the five classi-cal SE genes through multiplex-PCR.
Among the 60 isolates, 46 (76.7%) amplified femA: 41 were from RRM, four from mozzarella cheese and one from coalho cheese.
Identification of the Staphylococcal toxin encoding genes
Table 3 shows the results about toxin encoding genes. Among the 46 strains, 34 (74%) expressed one or more genes: 31 isolated from RRM and three from the mozzarella cheese. Twelve strains (26%) did not express any gene. All the toxin genes investigated (sea, seb, sec, sed, see and tst) were detected in the strains of S. aureus isolated from RRM. The three isolates from mozzarella cheese amplified only the gene responsible for the production of TSST-1; the sea gene was observed only in association with other genes, in contrast with the seb, sec, sed, see and tst genes, which were expressed separately.
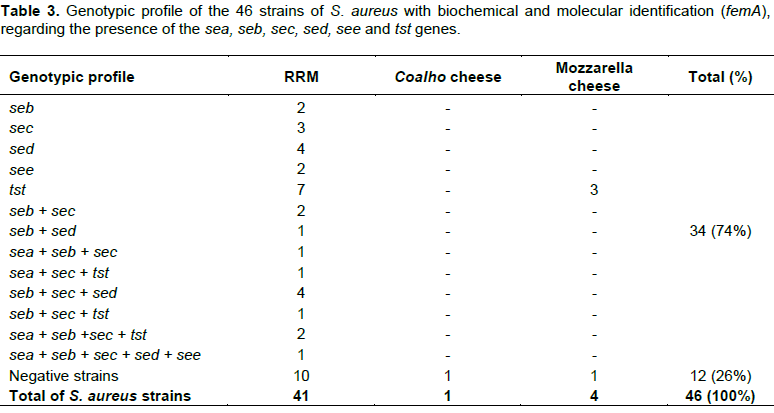
Thirteen different genotypes were obtained. The most frequent genotype was tst, which was present in 10 strains of S. aureus, of which seven were from RRM and three from mozzarella cheese. Twenty-one strains carrying a genotype with one toxin gene, three strains with two genes (seb + sec or seb + sed) and seven strains with three genes (sea + seb + sec; sea + sec + tst; seb + sec + sed; or seb + sec + tst) were observed.
The genotype encoding four toxins (sea + seb + sec + tst) was detected in two strains, and one strain of S. aureus expressed all five SE genes (sea + seb + sec + sed + see).
In Figure 2, the frequency of each of the genes investigated can be observed, independent of whether the expression was isolated or simultaneous. Out of all the genes investigated, sec was the most frequent one, observed in 15 strains (44.1%); followed by seb and tst, each in 14 (41.1%) strains; sed in 10 (29.4%), sea in five (14.7%) and see in three strains (8.8%) of S. aureus.
The highest percentage of S. aureus strains was isolated from the RRM samples. This was an expected result because all the samples presented high CPS contamination, with counts above 106 CFU/mL. This high CPS concentration could have occurred because this food is susceptible to contamination, particularly during the milking process, from the person performing the operation, from the utensils and equipment used, and even from one animal to another, especially in cases of mastitis in herds (Hait and Bennett, 2012).
Borges et al. (2008) also found that 100% of the RRM samples from a dairy in Ceará (Brazil) were contaminated by CPS with values between 103 and 106 CFU/mL. According to Sommerhäuser et al. (2003), the microbiological quality of milk is directly related to the hygiene of the milking process. The hygiene begins with the herd’s health, since many illnesses of dairy cattle affect the original composition, flavor, smell, viscosity and microbiological quality of the milk. Another aggravating factor is inadequate storage and temperature during transportation between the farm property and the dairy, which may contribute to multiplication of the contaminating microorganisms that were present at the time of the milking.
Although the law does not set limits for the presence of pathogenic microorganisms in RRM, according to FDA, in foods with CPS counts from 105 CFU/mL the presence of staphylococcal enterotoxin is likely (Hait and Bennett, 2012). Therefore, the counts measured in the present study could pose a great risk of presence of SEs in RRM, which could reach the cheese, even after the pasteurization process, which eliminates bacteria but does not destroy the toxins produced. The thermal stability of staphylococcal toxins favors endurance of these proteins in the thermal process, with the ability to withstand temperatures as high as 100ºC for 30 min, thus remaining active in foods (Balaban and Rasooly, 2000) and causing harm to human health.
S. aureus was not isolated in pasteurized milk and only a small number of samples presented CPS contamination, suggesting that the pasteurization process contributed to reduce the concentration of undesirable microbiota.
Although the pasteurization process ensures destruc-tion of the lineages of S. aureus that were originally present in the RRM, this bacterium may be found in PM if there is any flaw during the processing, leading to a cross-contamination and/or storage at inappropriate tem-perature (Corbia et al., 2000).
A study carried out in the state of São Paulo found that 38 (70.4%) out of 54 RRM samples presented CPS concentrations as high as 8.9 × 105 CFU/mL. There were eight PM samples with counts as high as 8.7 × 103 CFU/mL (Rall et al., 2008). Those values were lower than those found in the present study, which have found counts greater than 106 CFU/mL and 3.2 × 104 CFU/mL for RRM and PM, respectively.
The results from the cheese samples showed that 100% did not meet the standards required by Brazilian law (Brasil, 2001), which set limits for CPS in coalho and mozzarella cheese of up to 5×102 CFU/g and 103 CFU/g, respectively. Despite the low frequency of S. aureus isolation in cheese samples (coalho and mozzarella), high CPS concentrations pose a threat to public health because the production of toxins is not restricted only to the species S. aureus. Other CPS species can also produce toxins.
According to the International Commission on Microbiological Specifications for Foods (ICMSF, 1980), S. aureus counts between 103 and 104 CFU/g indicate a risk to public health. Values close to 105 CFU/g signify an epidemiological threat because of the possibility that enterotoxins might be present in quantities that are enough to cause staphylococcal intoxication, if the strain of S. aureus is toxigenic.
Post-pasteurization contamination occurs mainly due to inappropriate handling, lack of hygiene and deficient cleaning and sanitation of the equipment and utensils used in cheese production. Pelisser et al. (2009) highlighted that one of the main sources of CPS contamination in cheese are the handlers’ hands and forearms, due to deficient hygienic-sanitary control and no use of gloves during the processing.
Molecular characterization of Staphylococcus isolates showed that genetic analysis is more specific than biochemical tests in identifying this microorganism.
In a study carried out on dairy farms in various municipalities in the state of Minas Gerais, 100 strains of CPS were isolated. Among these, 77 were characterized as S. aureus by biochemical tests but 83 strains amplified the femA gene (Lange et al., 2011).
Several studies have explored the femA gene as a specific marker for S. aureus genotypic identification (Mehrotra et al., 2000, Riyaz-Ul-Hassan et al., 2008, Fischer et al., 2009, Pelisser et al., 2009), given that this gene takes part in biosynthesis of the pentaglycine interpeptide bridge that is characteristic for the peptidoglycan of the cell wall of this organism (Johnson et al., 1995, Moussallem et al., 2007).
Despite the high sensitivity of biochemical identification for characterizing S. aureus, its specificity is not 100% satisfactory. It needs to be complemented with molecular studies on specific markers for the microorganism.
Presence of the toxin encoding genes was observed in 74% (34) of the strains of S. aureus with biochemical and molecular identification. A great number of genotypes were found, divided in 13 different groups, thus indicating great genetic heterogeneity between the isolates.
Considering that in the present study only six toxin genes were investigated, it can be seen that the percentage of toxigenic S. aureus was high. This suggests that a great number of circulating strains of this pathogen carry toxin encoding genes. This would explain the high numbers of food poisoning cases and other infections commonly caused by this pathogen.
Studies carried out across the world have shown significant percentages of toxigenic S. aureus. In evaluating 78 strains of S. aureus isolated from milk from two farms in Tennessee with regard to the frequencies of 16 enterotoxin genes (sea-see and seg-seq) and the tst gene, it was observed that 73 strains (93.6%) carried one or more genes, comprising 36 different genotypic groups (Srinivasan et al., 2006).
In Italy, a study on 112 strains of S. aureus isolated from milk and dairy products found that 75 (67%) were positive for one or more SE genes (sea-see and seg-sel), divided into 17 genotypic profiles (Morandi et al., 2007).
Regarding the tst genotype, which was the one with highest frequency in the present study, Cardoso et al. (2000) and Zafalon et al. (2009) suggested that there might be a relationship between S. aureus strains carrying the tst gene and occurrences of cows with mastitis, and usually also in association with SE genes. In this, production of TSST-1 seemed to have great importance for the virulence of the samples of this microorganism, thereby influencing the severity of the cases of mastitis. In a study carried out in Brazil on 127 strains of S. aureus isolated from cases of clinical and subclinical mastitis, it was found that TSST-1 was one of the toxins produced with highest frequency. This was identified in 60% (475) of the samples, followed by SED (30%) and SEB (19%) (Silva et al., 2005). The presence of S. aureus carrying tst in refrigerated raw milk and in cheese could suggest that these isolates came from cows with mastitis, since TSST-1 has been associated with worsening of the inflammatory process of this illness among dairy cattle.
The strains of S. aureus that presented a genotypic profile with simultaneous presence of two to five toxin genes is a worrying finding because this shows the high pathogenic potential of these strains for production of different toxins, especially due to the high concentrations of the microorganism that were observed in all RRM and cheese samples.
In a study carried out in São Paulo on 132 strains of S. aureus isolates from raw milk, investigating the presence of the SE and TSST-1 genes, and the production of their respective toxins, 90 isolates (68.18%) were positive for one or two toxin genes, and 40 (44.44%) were capable of producing them in vitro (Chapaval et al., 2006).
Santana et al. (2010) reported that the risk of staphylococcal intoxication requires the presence of two factors: The food must contain staphylococci carrying the toxin genes with the ability to express this gene; and the microorganism counts should be higher than 105 CFU, under the conditions that allow toxin production in the food.
The presence of strains of S. aureus carrying toxigenic genes does not necessarily indicate production of toxins at levels sufficient to cause food poisoning conditions, because the production could be influenced by various factors (Le Loir et al., 2003, Hennekinne et al., 2012, BogdanoviÄová et al., 2017). However, the presence of these genes is required for the microorganism to be able to produce them. The PCR technique makes it possible to evaluate the genetic potential for such production and also serves as a screening test for confirming the presence of toxins in immunological assays (Zafalon et al., 2009).
Regarding the frequency of each gene investigated, the sec gene was the most prevalent, occurring in 15 strains (44.1%) of S. aureus, which was concordant with data from a study carried out in Germany, where 34 strains of S. aureus isolated from different dairy farms that amplified any gene (sea-see and tst) found that the sec gene occurred most frequently in 22 of the strains (64.7%), followed by the tst gene in 19 (55.8%) (Zschöck et al., 2000).
Divergent results were presented by Rall et al. (2008), who found that out of 57 strains of S. aureus isolated from raw and pasteurized milk, 39 (68.4%) were positive for at least one enterotoxin gene, among which the sea gene was the most frequent one, occurring in 16 strains (41%), followed by eight strains positive for sec (20.5%), five (12.8%) for sed, three for seb (7.7%) and two (5.1%) for see. Chapaval et al. (2006) also observed that the sea gene was the most frequent one in 90 strains of S. aureus, detected in 61 strains (67.78%), followed by tst in 38 (42.22%), seb in 30 (33.33%) and sec in five (5.56%), while no amplification of the sed and see genes occurred in any of the isolates.
The most frequently isolated staphylococcal enterotoxins from outbreaks of food poisoning are types A and D (Atanassova et al., 2001). In the United States, the enterotoxin A has been the type most involved, present in 77.8% of all outbreaks, followed by the enterotoxins D (37.5%) and B (10%) (Mathieu et al., 1991).
Enterotoxin types A and B have been associated with the contamination from food handlers, while types C and D have been correlated with animal-borne contamination, especially from cattle and pigs (Najera-Sanchez et al., 2003). Although outbreaks of staphylococcal intoxication have most commonly been attributed to ingestion of enterotoxin type A, and various studies have shown the prevalence of its respective gene, the data of the present study show that the sea gene was one of the least frequent ones, present only in five (14.7%) out of the 34 toxigenic strains of S. aureus.
The differences in occurrence of the toxin encoding genes between studies may be explained by the geogra-phic distribution and ecological origin of the strains, as well as by the sensitivity of the detection methods and the numbers and types of samples (Fagundes et al., 2010).
Based on our results, it can be concluded that all the genes of the classical enterotoxins (sea, seb, sec, sed and see) and the gene of the Toxic Shock Syndrome Toxin (tst) were identified in strains of toxigenic S. aureus, which presented high genetic heterogeneity and genetic potential for production of one or more toxins. All RRM and cheese samples from the dairies investigated presented high CPS counts. This suggests that the hygienic-sanitary quality was unsatisfactory and that a risk to public health could arise, due to the possible presence of toxins.
The authors have not declared any conflict of interests.
REFERENCES
|
Andreatta E, Fernandes AM, Santos MV, Mussarelli C, Marques MC, Oliveira CAF (2009). Composition, functional properties and sensory characteristics of Mozzarella cheese. Braz. Arch. Biol. Technol. 52(5):1235-1242.
Crossref
|
|
|
|
Atanassova V, Meindl A, Ring C (2001). Prevalence of Staphylococcus aureus and staphylococcal enterotoxins in raw pork and uncooked smoked ham--a comparison of classical culturing detection and RFLP-PCR. Int. J. Food Microbiol. 68(1-2):105-113.
Crossref
|
|
|
|
|
Balaban N, Rasooly A (2000). Staphylococcal enterotoxins. Int. J. Food Microbiol. 61(1):1-10.
Crossref
|
|
|
|
|
Becker K, Roth R, Peters G (1998). Rapid and specific detection of toxigenic Staphylococcus aureus: use of two multiplex PCR enzyme immunoassays for amplification and hybridization of staphylococcal enterotoxin genes, exfoliative toxin genes, and toxic shock syndrome toxin 1 gene. J. Clin. Microbiol. 36(9):2548-2553.
|
|
|
|
|
BogdanoviÄová K, Necidová L, Haruštiaková D, Janštová B (2017). Milk powder risk assessment with Staphylococcus aureus toxigenic strains. Food Contr. 73:2-7.
Crossref
|
|
|
|
|
Borges MF, Nassu RT, Pereira JL, Andrade APC, Kuaye AY (2008). Perfil de contaminação por Staphylococcus e suas enterotoxinas e monitorização das condições de higiene em uma linha de produção de queijo de coalho. Cien. Rural. 38(5):1431-1438.
Crossref
|
|
|
|
|
Brasil (2001). ANVISA - Agência Nacional de Vigilância Sanitária. Resolução RDC nº 12, de 02 de janeiro de 2001.
|
|
|
|
|
Cardoso HFT, Carmo LS, Silva N (2000). Detecção da toxina-1 da síndrome do choque tóxico em amostras de Staphylococcus aureus isoladas de mastite bovina. Arq. Bras. Med. Vet. Zootec. 52(1):7-10.
Crossref
|
|
|
|
|
Chapaval L, Moon DH, Gomes JE, Duarte FR, Tsai SM (2006). Use of PCR to detect classical enterotoxins genes (ent) and toxic shock syndrome toxin-1 gene (tst) in Staphylococcus aureus isolated from crude milk and determination of toxin productivities of S. aureus isolates harboring this genes. Arq. Inst. Biol. 73(2):165-169.
|
|
|
|
|
Corbia ACG, Nascimento MGF, Oliveira CZF, Nascimento ER (2000). Staphylococcus aureus: importância para a saúde pública e aspectos epidemiológicos. Embrapa Agrobiologia, Seropédica. 114:1-15.
|
|
|
|
|
Fagundes H, Barchesi L, Filho AN, Ferreira LM, Oliveira CAF (2010). Occurrence of Staphylococcus aureus in raw milk produced in dairy farms in São Paulo state, Brazil. Braz. J. Microbiol. 41(2):376-380.
Crossref
|
|
|
|
|
Fischer A, Francois P, Holtfreter S, Broeker B, Schrenzel J (2009). Development and evaluation of a rapid strategy to determine enterotoxin gene content in Staphylococcus aureus. J. Microbiol. Methods. 77(2):184-190.
Crossref
|
|
|
|
|
Garcia LS (2010). Biochemical Tests for the Identification of Aerobic Bacteria. Pages 503-642 in Clinical Microbiology Procedures Handbook. 3rd ed. ASM Press, Washington, DC, USA.
Crossref
|
|
|
|
|
Guimaraes FF, Nobrega DB, Richini-Pereira VB, Marson PM, Figueiredo Pantoja JC, Langoni H (2013). Enterotoxin genes in coagulase-negative and coagulase-positive staphylococci isolated from bovine milk. J. Dairy Sci. 96(5):2866-2872.
Crossref
|
|
|
|
|
Hait J, Bennett R (2012). Staphylococcus aureus. in Bad Bug Book, Foodborne Pathogenic Microorganisms and Natural Toxins. 2 ed. Lampel KA, Al-Khaldi S, and Cahill SM, ed. Center for Food Safety and Applied Nutrition (CFSAN) of the Food and Drug Administration (FDA).
|
|
|
|
|
Haug A, Hostmark AT, Harstad OM (2007). Bovine milk in human nutrition--a review. Lipids Health Dis. 6:25.
Crossref
|
|
|
|
|
Hennekinne JA, De Buyser ML, Dragacci S (2012). Staphylococcus aureus and its food poisoning toxins: characterization and outbreak investigation. FEMS Microbiol. Rev. 36(4):815-836.
Crossref
|
|
|
|
|
ICMSF (1980). International Comission on Microbiological Specifications for Foods. Ecologia microbiana de los alimentos. Pages 1-38. Acribia, Zaragoza.
|
|
|
|
|
Johnson S, Kruger D, Labischinski H (1995). FemA of Staphylococcus aureus: isolation and immunodetection. FEMS Microbiol. Lett. 132(3):221-228.
Crossref
|
|
|
|
|
Kadariya J, Smith TC, Thapaliya D (2014). Staphylococcus aureus and staphylococcal food-borne disease: an ongoing challenge in public health. Biomed. Res. Int. 9p.
|
|
|
|
|
Lange CC, Brito MAVP, Brito JRF, Arcuri EF, Souza GN, Machado MA, Domingues R, Salimena APS (2011). Uso de PCR e sequenciamento do rDNA 16S para identificação de bactérias do gênero Staphylococcus isoladas de mastite bovina. Pesq. Vet. Bras. 31(1):36-40.
Crossref
|
|
|
|
|
Le Loir Y, Baron F, Gautier M (2003). Staphylococcus aureus and food poisoning. Genet. Mol. Res. 2(1):63-76.
|
|
|
|
|
Martin JG, de Oliveira ESG, da Fonseca CR, Morales CB, Souza Pamplona Silva C, Miquelluti DL, Porto E (2016). Efficiency of a cleaning protocol for the removal of enterotoxigenic Staphylococcus aureus strains in dairy plants. Int. J. Food Microbiol. 238:295-301.
Crossref
|
|
|
|
|
Mathieu AM, Isigidi BK, Devriese LA, Godard C, Vanhoof R (1991). Characterization of Staphylococcus aureus and Salmonella spp. strains isolated from bovine meat in Zaire. Int. J. Food Microbiol. 14(2):119-125.
Crossref
|
|
|
|
|
Mehrotra M, Wang G, Johnson WM (2000). Multiplex PCR for detection of genes for Staphylococcus aureus enterotoxins, exfoliative toxins, toxic shock syndrome toxin 1, and methicillin resistance. J. Clin. Microbiol. 38(3):1032-1035.
|
|
|
|
|
Moraes PM, Vicosa GN, Yamazi AK, Ortolani MB, Nero LA (2009). Foodborne pathogens and microbiological characteristics of raw milk soft cheese produced and on retail sale in Brazil. Foodborne Pathog. Dis. 6(2):245-249.
Crossref
|
|
|
|
|
Morandi S, Brasca M, Lodi R, Cremonesi P, Castiglioni B (2007). Detection of classical enterotoxins and identification of enterotoxin genes in Staphylococcus aureus from milk and dairy products. Vet. Microbiol. 124(1-2):66-72.
Crossref
|
|
|
|
|
Moussallem BC, Kury CMH, Medina-Acosta E (2007). Detecção dos genes mecA e femA, marcadores moleculares de resistência a meticilina, em Staphylococcus spp. isolados de pacientes admitidos em uma Unidade Neonatal de Tratamento Intensivo. Rev. Cient. Fac. Med. Campos. 2:2-9.
|
|
|
|
|
Najera-Sanchez G, Maldonado-Rodriguez R, Ruiz Olvera P, de la Garza LM (2003). Development of two multiplex polymerase chain reactions for the detection of enterotoxigenic strains of Staphylococcus aureus isolated from foods. J. Food Prot. 66(6):1055-1062.
Crossref
|
|
|
|
|
Nunes MM, Caldas ED (2017). Preliminary Quantitative Microbial Risk Assessment for Staphylococcus enterotoxins in fresh Minas cheese, a popular food in Brazil. Food Contr. 73:524-531.
Crossref
|
|
|
|
|
Oliver SP, Jayarao BM, Almeida RA (2005). Foodborne pathogens in milk and the dairy farm environment: food safety and public health implications. Foodborne Pathog. Dis. 2(2):115-129.
Crossref
|
|
|
|
|
Ortega E, Abriouel H, Lucas R, Galvez A (2010). Multiple roles of Staphylococcus aureus enterotoxins: pathogenicity, superantigenic activity, and correlation to antibiotic resistance. Toxins (Basel). 2(8):2117-2131.
Crossref
|
|
|
|
|
Pelisser MR, Klein CS, Ascoli KR, Zotti TR, Arisi ACM (2009). Ocurrence of Staphylococcus aureus and multiplex pcr detection of classic enterotoxin genes in cheese and meat products. Braz. J. Microbiol. 40(1):145-148.
Crossref
|
|
|
|
|
Rall VL, Vieira FP, Rall R, Vieitis RL, Fernandes A, Jr., Candeias JM, Cardoso KF, Araujo JP (2008). PCR detection of staphylococcal enterotoxin genes in Staphylococcus aureus strains isolated from raw and pasteurized milk. Vet. Microbiol. 132(3-4):408-413.
Crossref
|
|
|
|
|
Riyaz-Ul-Hassan S, Verma V, Qazi GN (2008). Evaluation of three different molecular markers for the detection of Staphylococcus aureus by polymerase chain reaction. Food Microbiol. 25(3):452-459.
Crossref
|
|
|
|
|
Santana EHW, Cunha MLRS, Oliveira TCRMd, Moraes LB, Aragon-Alegro LC, Beloti V (2010). Assessment of the risk of raw milk consumption related to staphylococcal food poisoning. Cien. Anim. Bras. 11(3):643-652.
|
|
|
|
|
Scharff RL, Besser J, Sharp DJ, Jones TF, Peter GS, Hedberg CW (2016). An Economic Evaluation of PulseNet: A Network for Foodborne Disease Surveillance. Am. J. Prev. Med. 50(5 Suppl 1):S66-73.
Crossref
|
|
|
|
|
Silva ER, Carmo LS, Silva N (2005). Detection of the enterotoxins A, B, and C genes in Staphylococcus aureus from goat and bovine mastitis in Brazilian dairy herds. Vet. Microbiol. 106(1-2):103-107.
Crossref
|
|
|
|
|
Silva RA, Lima MS, Viana JB, Bezerra VS, Pimentel MC, Porto AL, Cavalcanti MT, Lima Filho JL (2012). Can artisanal "Coalho" cheese from Northeastern Brazil be used as a functional food? Food Chem. 135(3):1533-1538.
Crossref
|
|
|
|
|
Sommerhäuser J, Kloppert B, Wolter W, Zschöck M, Sobiraj A, Failing K (2003). The epidemiology of Staphylococcus aureus infections from subclinical mastitis in dairy cows during a control programme. Vet. Microbiol. 96(1):91-102.
Crossref
|
|
|
|
|
Srinivasan V, Sawant AA, Gillespie BE, Headrick SJ, Ceasaris L, Oliver SP (2006). Prevalence of enterotoxin and toxic shock syndrome toxin genes in Staphylococcus aureus isolated from milk of cows with mastitis. Foodborne Pathog. Dis. 3(3):274-283.
Crossref
|
|
|
|
|
Zafalon LF, Arcaro JRP, Filho AN, Ferreira LM, Veschi JLA (2009). Staphylococcus aureus portadores de genes de toxinas isolados em amostras de diferentes fontes de transmissão durante a ordenha Rev. Inst. Adolfo Lutz. 68(2):269-277.
|
|
|
|
|
Zschöck M, Botzler D, Blöcher S, Sommerhäuser J, Hamann HP (2000). Detection of genes for enterotoxins (ent) and toxic shock syndrome toxin-1 (tst) in mammary isolates of Staphylococcus aureus by polymerase-chain-reaction. Int. Dairy J. 10:569-574.
Crossref
|
|