Full Length Research Paper
ABSTRACT
Mangrove ecosystems provide a unique ecological niche for diverse microbial communities. This study aimed to identify bacterial isolates from the rhizospheres of four mangrove species (Sonneratia alba, Rhizophora mucronata, Ceriops tagal and Avicennia marina) using the 16S rRNA gene analysis approach. Rhizospheric sediment samples of the mangroves were collected from Mida creek and Gazi bay, Kenya, using standard protocols. A total of 36 representative bacterial isolates were analyzed. The isolates were characterized using morphological and molecular characters. Pure gDNA was extracted from the isolates, polymerase chain reaction amplified and sequenced. The 16S rRNA gene sequences were BLASTN analyzed against the Genbank database; the closest taxonomically related bacterial sequences were retrieved and used for phylogenetic analysis using MEGA X software. Morphologically, the isolates differed in their cultural characteristic in color, shape, margin, elevation and gram reaction. Phylogenetic analysis classified the isolates into five genera, namely Bacillus, Pseudomonas, Micrococcus, Microbacterium and Streptomyces that belong to three different phyla (Firmicutes, Proteobacteria and Actinobacteria). The findings show that the underexplored tropical mangrove rhizospheres harbor useful diverse bacteria. Further analysis of the bioactive production potential of the isolates will give more insights into the types of bioactive compounds produced and their biotechnological potential.
Key words: 16S rRNA gene sequence, rhizosphere, mangrove sediments, marine bacteria, biotechnology.
INTRODUCTION
MATERIALS AND METHODS
RESULTS
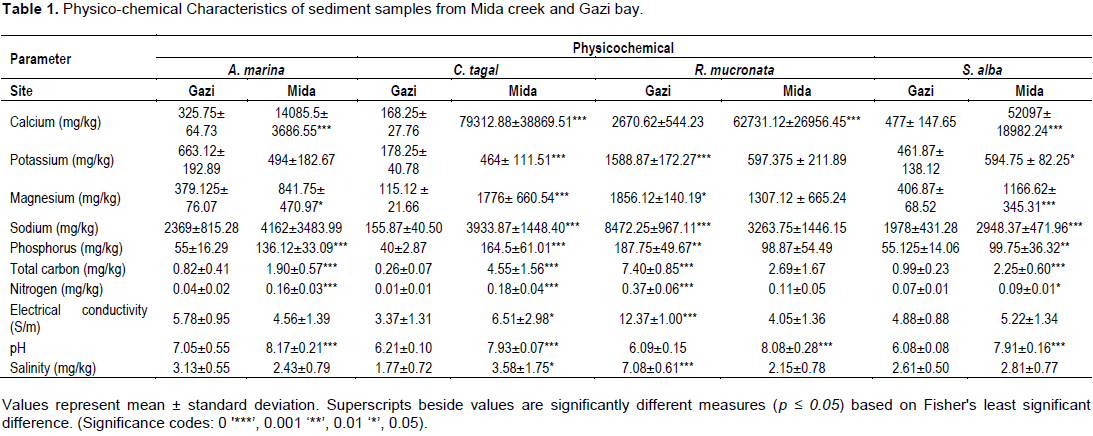
DISCUSSION
CONFLICT OF INTERESTS
ACKNOWLEDGEMENTS
REFERENCES
|
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990). Basic local alignment search tool. Journal of Molecular Biology 215(3):403-410. |
|
|
Anna PSAR, Bruno FRdeO, Renan de SS, Igor DAR, Ariana AR, Danns PB, Matheus MV, José DGV (2018). Isolation and characterization of phenol degrading Bacillus species from Southeast Brazilian mangrove sediment. African Journal of Microbiology Research 12(46):1032-1038. |
|
|
Ashelford KE, Chuzhanova NA, Fry JC, Jones AJ, Weightman AJ (2006). New screening software shows that most recent large 16S rRNA gene clone libraries contain chimeras. Applied and Environmental Microbiology 72(9):5734-5741. |
|
|
Azman AS, Othman I, Velu SS, Chan KG, Lee LH (2015). Mangrove rare actinobacteria: Taxonomy, natural compound, and discovery of bioactivity. Frontiers in Microbiology 6:1-15. |
|
|
Barka EA, Vatsa P, Sanchez L, Gaveau-vaillant N, Jacquard C, Klenk H, Clément C, Ouhdouch Y, Wezel P (2016). Taxonomy, Physiology, and Natural Products of Actinobacteria. Microbiology and Molecular Biology Reviews 80(1):1-44. |
|
|
Behera BC, Parida S, Dutta SK, Thatoi HN (2014a). Isolation and Identification of Cellulose Degrading Bacteria from Mangrove Soil of Mahanadi River Delta and Their Cellulase Production Ability. American Journal of Microbiological Research 2(1):41-46. |
|
|
Behera BC, Patra M, Dutta SK, Thatoi HN (2014b). Isolation and Characterization of Sulphur Oxidising Bacteria from Mangrove Soil of Mahanadi River Delta and Their Sulphur Oxidising Ability. Journal of Applied and Environmental Microbiology 2(1):1-5. |
|
|
Bergey DH, Holt JG (1994). Bergey's manual of determinative bacteriology. |
|
|
Brupbacher RH, Bonner WP, Sedberry JRJ (1968). Analytical methods and procedures used in the soil testing laboratory: 15. View |
|
|
Bull AT, Ward AC, Goodfellow M (2000). Search and Discovery Strategies for Biotechnology: The Paradigm Shift. Microbiology and Molecular Biology Reviews 64(3):573-606. |
|
|
Clarridge JE (2004). Impact of 16S rRNA Gene Sequence Analysis for Identification of Bacteria on Clinical Microbiology and Infectious Diseases. Clinical Microbiology Reviews 17(4):840-862. |
|
|
Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarrell DM, Marsh T, Garrity GM, Tiedje JM (2009). The Ribosomal Database Project: Improved alignments and new tools for rRNA analysis. Nucleic Acids Research 37(1):141-145. |
|
|
Conlan S, Kong HH, Segre JA (2012). Species-Level Analysis of DNA Sequence Data from the NIH Human Microbiome Project. PLoS ONE 7(10). |
|
|
Dahdouh-Guebas F, Van Pottelbergh I, Kairo JG, Cannicci S, Koedam N (2004). Human-impacted mangroves in Gazi (Kenya): Predicting future vegetation based on retrospective remote sensing, social surveys, and tree distribution. Marine Ecology Progress Series 272:77-92. |
|
|
de Mendiburu F (2020). agricolae: Statistical Procedures for Agricultural Research. R package version 1.3-2. |
|
|
Dissanayake N, Chandrasekara U (2014). Effects of Mangrove Zonation and the Physicochemical Parameters of Soil on the Distribution of Macrobenthic Fauna in Kadolkele Mangrove Forest, a Tropical Mangrove Forest in Sri Lanka. Advances in Ecology, pp. 1-13. |
|
|
Felsenstein J (1985). Confidence Limits on Phylogenies: An Approach Using the Bootstrap. Evolution 39(4):783. |
|
|
Fettweis JM, Serrano MG, Sheth NU, Mayer CM, Glascock AL, Brooks JP, Jefferson KK, Buck GA (2012). Species-level classification of the vaginal microbiome. BMC Genomics 13(8):1-9. |
|
|
Giannopoulos G, Lee DY, Neubauer SC, Brown BL, Franklin RB (2019). A simple and effective sampler to collect undisturbed cores from tidal marshes. |
|
|
Goessens A, Satyanarayana B, Van Der Stocken T, Zuniga MQ, Mohd-Lokman H, Sulong I, Dahdouh-Guebas F (2014). Is Matang Mangrove Forest in Malaysia sustainably rejuvenating after more than a century of conservation and harvesting management? PLoS ONE 9:8. |
|
|
Haldar S, Nazareth SW (2018). Taxonomic diversity of bacteria from mangrove sediments of Goa: metagenomic and functional analysis. 3 Biotech 8:436-446. |
|
|
Holguin G, Vazquez P, Bashan Y (2001). The role of sediment microorganisms in the productivity, conservation, and rehabilitation of mangrove ecosystems: An overview. Biology and Fertility of Soils 33(4):265-278. |
|
|
Jenoh EM, De Villiers EP, De Villiers SM, Okoth S, Jefwa J, Kioko E, Kaimenyi D, Hendrickx M, Dahdouh-Guebas F, Koedam N (2019). Infestation mechanisms of two woodborer species in the mangrove Sonneratia alba J. Smith in Kenya and co-occurring endophytic fungi. PLoS ONE 14(10):1-20. |
|
|
Jenoh EM, Robert EMR, Lehmann I, Kioko E, Bosire JO, Ngisiange N, Dahdouh-Guebas F, Koedam N (2016). Wide ranging insect infestation of the pioneer mangrove Sonneratia alba by two insect species along the Kenyan coast. PLoS ONE 11(5):1-15. |
|
|
Josiah OK, Huxley MM, Hamadi IB, Anne TWM, Jun U (2018). Phylogenetic diversity of prokaryotes on the snow-cover of Lewis glacier in Mount Kenya. African Journal of Microbiology Research 12(24):574-579. |
|
|
Kambura AK, Mwirichia RK, Kasili RW, Karanja EN, Makonde HM, Boga HI (2016a). Bacteria and Archaea diversity within the hot springs of Lake Magadi and Little Magadi in Kenya. BMC Microbiology 16(1):1-12. |
|
|
Kambura AK, Romano KM, Remmy WK, Edward NK, Huxley MM, Hamadi IB (2016b). Diversity of fungi in sediments and water sampled from the hot springs of Lake Magadi and Little Magadi in Kenya. African Journal of Microbiology Research 10(10):330-338. |
|
|
Kathiresan K, Selvam MM (2006). Evaluation of beneficial bacteria from mangrove soil. Botanica Marina 49(1):86-88. |
|
|
Kawaka F, H, Dida M, Opala P, Ombori O, Maingi J, Muoma J (2018). Genetic diversity of symbiotic bacteria nodulating common bean (Phaseolus vulgaris) in western Kenya. PLoS ONE 13(11):1-13. |
|
|
Krüger C, Kohout P, Janoušková M, Püschel D, Frouz J, Rydlová J (2017). Plant communities rather than soil properties structure arbuscular mycorrhizal fungal communities along primary succession on a mine spoil. Frontiers in Microbiology 8:1-1. |
|
|
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018). MEGA X: Molecular evolutionary genetics analysis across computing platforms. Molecular Biology and Evolution 35(6):1547-1549. |
|
|
Kunasundari B, Naresh S, Che Zakaria NZ (2017). Isolation and characterization of cellulase producing bacteria from tropical mangrove soil. ACM International Conference Proceeding Series Part F131935(9):34-37. |
|
|
Kurniawan A, Prihanto AA, Sari SP, Febriyanti D, Kurniawan A, Sambah AB, Asriani E (2018). Isolation and Identification of cellulolytic bacteria from mangrove sediment in Bangka Island. IOP Conference Series: Earth and Environmental Science 137(1):0-6. |
|
|
Ladeira SA, Cruz E, Delatorre AB, Barbosa JB, Martins MLL (2015). Cellulase production by thermophilic Bacillus sp. SMIA-2 and its detergent compatibility. Electronic Journal of Biotechnology 18(2):110-115. |
|
|
Lang'at JKS (2008). Variability of mangrove forests along the Kenyan coast. |
|
|
Lee L, Zainal N, Azman A, Eng S, Goh B, Yin W, Mutalib NA, Chan K (2014). Diversity and Antimicrobial Activities of Actinobacteria Isolated from Tropical Mangrove Sediments in Malaysia. Hindawi: 1-14. |
|
|
Makonde HM, Mwirichia R, Osiemo Z, Boga HI, Klenk HP (2015). 454 Pyrosequencing-based assessment of bacterial diversity and community structure in termite guts, mounds and surrounding soils. SpringerPlus 4(1):471. |
|
|
Maldonado LA, Stach JEM, Pathom-Aree W, Ward AC, Bull AT, Goodfellow M (2005). Diversity of cultivable actinobacteria in geographically widespread marine sediments. International Journal of General and Molecular Microbiology 87(1):11-18. |
|
|
Malek NA, Jalal A, Chowdhury K, Zainuddin Z (2014). Selective Isolation of Actinomycetes from Mangrove Forest of Pahang, Malaysia. International Conference on Agriculture, Biology and Environmental Sciences 14:9-13. |
|
|
Matthijs S, Tack J, van Speybroeck D, Koedam N (1999). Mangrove species zonation and soil redox state, sulphide concentration and salinity in Gazi Bay (Kenya), a preliminary study. Mangroves and Salt Marshes 3(4):243-249. |
|
|
Mendes R, Garbeva P, Raaijmakers JM (2013). The rhizosphere microbiome: Significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiology Reviews 37(5):634-663. |
|
|
Mo K, Huang H, Bao S, Hu Y (2020). Bacillus caeni sp. Nov., isolated from mangrove sediment. International Journal of Systematic and Evolutionary Microbiology 70(3):1503-1507. |
|
|
Mohamed MOS, Neukermans G, Kairo JG, Dahdouh-Guebas F, Koedam N (2009). Mangrove forests in a peri-urban setting: The case of Mombasa (Kenya). Wetlands Ecology and Management 17(3):243-255. |
|
|
Mohan YSYVJ, Sirisha B, Prathyusha K, Rao P (2014). Isolation, Screening and Characterization of Actinomycetes from Marine Sediments for their Potential to Produce Antifungal Agents. International Journal of Life Sciences, Biotechnology and Pharmaceutical Research 3(4):131-137. |
|
|
Muhonja CN, Magoma G, Imbuga M, Makonde HM (2018a). Molecular characterization of Low-Density Polyethene (LDPE) degrading bacteria and fungi from Dandora dumpsite, Nairobi, Kenya. International Journal of Microbiology. Article ID 4167845. |
|
|
Muhonja CN, Makonde H, Magoma G, Imbuga M (2018b). Biodegradability of polyethylene by bacteria and fungi from Dandora dumpsite Nairobi-Kenya. PLoS ONE 13(7):1-17. |
|
|
Muwawa EM, Nancy LMB, Zipporah LO, Hamadi IB, Huxley MM (2016). Isolation and characterization of some gut microbial symbionts from fungus-cultivating termites (Macrotermes and Odontotermes spp.). African Journal of Microbiology Research 10(26):994-1004. |
|
|
Naik G, Shukla S, Kumar MS (2013). Isolation and Characterization of Actinomycetes Isolates for Production of Antimicrobial Compounds. Journal of Microbiology and Biotechnology Research 3(5):33-36. |
|
|
Naresh S, Kunasundari B, Gunny AAN, Teoh YP, Shuit SH, Ng QH, Hoo PY (2019). Isolation and partial characterisation of thermophilic cellulolytic bacteria from north Malaysian tropical mangrove soil. Tropical Life Sciences Research 30(1):123-147. |
|
|
Ntabo RM, Nyamache AK, Lwande W, Kabii J, Nonoh J (2018). Enzymatic Activity of Endophytic Bacterial Isolates from Selected Mangrove Plants in Kenya. The Open Microbiology Journal 12(1):354-363. |
|
|
Priya E, Thenmozhi R, Nagasathya A, Thajuddin N, Muralitharan G (2014). Diversity of Actinobacteria in Mangrove Ecosystem of Muthupet, India. International Research Journal of Environment Sciences 3(4):13-17. |
|
|
Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, Glöckner FO (2007). SILVA: A comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Research 35(21):7188-7196. |
|
|
Pupin B, Nahas E (2014). Microbial populations and activities of mangrove, restinga and Atlantic forest soils from Cardoso Island, Brazil. Journal of Applied Microbiology 116(4):851-864. |
|
|
Rasigraf O, Helmond NAGM, van Frank J, Lenstra WK, Egger M, Slomp C.P, Jetten MSM (2019). Metagenomic analysis reveals large potential for carbon, nitrogen and sulfur cycling in coastal methanic sediments of the Bothnian Sea. BioRxiv-Pre-Print: 553131. |
|
|
Saitou N, Nei M (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molecular Biology and Evolution 4(4):406-425. |
|
|
Salano OA, Huxley MM, Remmy WK, Hamadi IB (2018). Isolation and characterization of fungi from a hot-spring on the shores of Lake Bogoria, Kenya. Journal of Yeast and Fungal Research 9(1):1-13. |
|
|
Salano OA, Makonde HM, Kasili RW, Wangai LN, Nawiri MP, Boga HI (2017). Diversity and distribution of fungal communities within the hot springs of soda lakes in the Kenyan rift valley. African Journal of Microbiology Research 11(19):764-775. |
|
|
Sanders CJ, Eyre BD, Santos IR, Machado W, Luiz-silva W, Smoak JM, Breithaupt JL, Ketterer ME, Sanders L, Marotta H, Silva-filho E, Al SET (2014). Impacted Mangrove Wetland. Geophysical Research Letters 41(1):2475-2480. |
|
|
Sarker A, Haque M, Islam M, Rahman M, Islam M (2015). Isolation and Characterization of a Marine Bacterium from Sundarbans, Bangladesh. British Microbiology Research Journal 6(6):348-357. |
|
|
Saseeswari A, Kanimozhi G, Panneerselvam A (2016). Bacterial Diversity of Mangrove Soil in Karankadu from East Coast of Tamil Nadu, India. International Journal of Current Microbiology and Applied Sciences 5(4):750-756. |
|
|
Sengupta S, Pramanik A, Ghosh A, Bhattacharyya M (2015). Antimicrobial activities of actinomycetes isolated from unexplored regions of Sundarbans mangrove ecosystem. BMC Microbiology 15(1):1-16. |
|
|
Soares-Júnior FL, Dias ACF, Fasanella CC, Taketani RG, Lima AOdeS, Melo IS, Andreote FD (2013). Endo-and exoglucanase activities in bacteria from mangrove sediment. Brazilian Journal of Microbiology 44(3):969-976. |
|
|
Somanathan H, Mali S, Borges RM, Shilton LA, Altringham JD, Compton SG, Whittaker RJ, Russo SE, Augspurger CK, Nakashima Y, Inoue E, Inoue-Murayama M, Sukor JR, McConkey KR, Prasad S, Corlett RT, Campos-Arceiz A, Brodie JF, Rogers H, Rawat GS (2004). R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing. Oecologia 15:413-448. |
|
|
Srinivasan R, Karaoz U, Volegova M, MacKichan J, Kato-Maeda M, Miller S, Nadarajan R, Brodie EL, Lynch SV (2015). Use of 16S rRNA gene for identification of a broad range of clinically relevant bacterial pathogens. PLoS ONE 10(2):1-22. |
|
|
Tam HT, City CT, Diep CN, City CT (2017). Isolation and Characterization of Bacteria of Mangrove Rhizosphere in the Mekong Delta, Vietnam. International Journal of Innovations in Engineering and Technology 9(1):68-79. |
|
|
Tamura K, Nei M, Kumar S (2004). Prospects for inferring very large phylogenies by using the neighbor-joining method. Proceedings of the National Academy of Sciences of the United States of America 101(30):11030-11035. |
|
|
Wang Q, Garrity GM, Tiedje JM, Cole JR (2007). Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Applied and Environmental Microbiology 73(16):5261-5267. |
|
|
Wu P, Xiong X, Xu Z, Lu C, Cheng H, Lyu X, Zhang J, He W, Deng W, Lyu Y, Lou Q, Hong Y, Fang H (2016). Bacterial communities in the rhizospheres of three mangrove tree species from Beilun Estuary, China. PLoS ONE 11(10):1-13. |
|
|
Xu DB, Ye WW, Han Y, Deng ZX, Hong K (2014). Natural products from mangrove actinomycetes. Marine Drugs 12(5):2590-2613. |
|
|
Zhang Y, Yang Q, Ling J, Van Nostrand JD, Shi Z, Zhou J, Dong J (2017). Diversity and structure of diazotrophic communities in mangrove rhizosphere, revealed by high-throughput sequencing. Frontiers in Microbiology 8(8):1-11. |
|
Copyright © 2024 Author(s) retain the copyright of this article.
This article is published under the terms of the Creative Commons Attribution License 4.0