Full Length Research Paper
ABSTRACT
This study determined the prevalence and drug resistant patterns of bacteria isolated from untreated hospital wastewaters collected from selected hospitals in Offa Local Government Area of Kwara State, Nigeria. A total of 42 composite samples were aseptically collected, transported and analyzed for enumeration of microorganisms, bacteriological identification and susceptibility testing following standard procedures. The Global Positioning System (GPS) coordinates of each site location was equally taken and data obtained were analyzed using SPSS version 20. The means bacterial count population of wet season samples ranged between 7±4.00 × 105 and 150±43.59 × (105cfu/ml), while that of dry season samples ranged between 10±2.00 × 105 and 225±67.27 × 105 cfu/ml. Among the total samples, 50 bacterial isolates were detected, of which 26(52%) were from wet season samples and 24(48%) were from dry season samples. The most frequently isolated bacteria from wet season samples was Alcaligenes faecalis 17(65.4%) followed by Alcaligenes aquatilis 5(19.2%) and Staphylococcus saprophyticus 4(15.4%). Findings from antibiotic resistance pattern of the isolates indicated that ofloxacin (OFL) demonstrated highest antimicrobial potency against the test isolates, with Zone inhibition diameters (mm) (resistant ≤12, intermediate 13-15 and susceptible ≥ 16). Thus, hospital wastewater should be treated before discharge to prevent infectious diseases.
Key words: Hospital wastewaters, antibiotics, resistant pattern, bacteria.
INTRODUCTION
Wastewater is any water that has had its quality severely degraded by human intervention. This comprises liquid waste from private residences, businesses, industries, hospitals, and agricultural and commercial establishments (Verlicchi et al., 2010). Patient wards, surgery units laboratories, clinical wards, laundries, and other areas of a hospital generate wastewater, which has a wide range of loads based on the activities carried out (Aurelien
et al., 2013).
Owing to several difficulties, hospital effluent has received a lot of attention in the last few years in many nations throughout the world. Hospitals are known to consume large amounts of water each day, ranging from 400 to 1200 L per day, resulting in a comparable volume of water burden (Gautam et al., 2007).
Hospital wastewater is an ideal medium for microorganisms and carries the resistant gene into the sewage system (Abdel-Rouf et al., 2012; Fekadu et al., 2015). Pharmaceuticals, radionuclides, detergent, antibiotics, antiseptic, surfactant, solvent, medicinal medication, heavy metals, and radioactive substances are among the persistent chemical compounds and complex combinations of organic matter found in hospital wastewater (Aurelien et al., 2013; Ferrando-Climent et al., 2014) plus microorganisms such as Bacillus subtilis, Staphylococcus aureus, Klebsiella pneumonia and Escherichia coli (Anitha and Jayraaj, 2012; Ferrando-Climent et al., 2014).
Medicines are used in large quantities in hospitals on a daily basis for patient care and infection control, and a significant quantity of these antibiotics is excreted through the faeces and urine of patients, eventually reaching liquid wastes. As a result, hospital wastewater contains resistant gene and antibiotic residues, which, through selection pressure, hinders the growth of vulnerable microorganisms (Beyene and Redaie, 2011; Stalder et al., 2014). Additionally, some of these substances and bacteria defecated by patients are detected in hospital wastewater, which is then discharged into the local sewer system without being treated. As a result, this composition causes a wide range of toxicity, genotoxicity, and organic load, resulting in a completely negative influence on the natural ecosystem and an inherent health threat to humans (Wilde et al., 2013).
By functioning as a vector or reservoir of resistant gene, resistant bacteria in the environment contain transmissible gene (Pandey et al., 2011; Keen and Patrick, 2013). They are also the most hazardous microorganisms that pose a health risk to human, and wastewaters are one of the most serious contaminants that pollute the environment (Diwan et al., 2010; Pandey et al., 2011).
Antibiotic resistance is becoming a major public health issue around the world. The World Health Organization (WHO) and the European Commission (EC) have both acknowledged the necessity of researching the genesis and spread of resistance, as well as the need for control tactics (Oteo et al., 2001). In developing countries like Nigeria, improper antibiotic use, ineffective infection control programs, and a lack of better management of hospital wastewater are the main factors for antimicrobial resistance gene dissemination in the environment. Therefore, this study aimed to exploit the role of hospital wastewater as source of emerging drug resistance pathogens in the environment. The objectives of this research were to isolate bacteria associated with some hospital wastewaters and determine their antibiotics resistant patterns.
MATERIALS AND METHODS
Study area
Offa Local Government is one of the major Local Government Areas and one of the major cities of Kwara State, situated in the North Central geographical zone of Nigeria. It is situated between latitude 8° 10' 33" N, 238 km North of Lagos at longitude 4° 43' 02" E and 530 km South West of Abuja; the Federal Capital of Nigeria (ODUNA, 2020). The yearly temperature ranges between 18.5 - 37.5°C, with a humidity of between 45 – 47%, and a peak annual rainfall of 200 cm (Wikipedia, 2020). The vegetation is essentially guinea savanna, a transition zone between the Sudan savanna of the far North and the tropical rain forest vegetation of the South East. The population of Offa from the 2006 census was estimated at 88,975, while the annual growth rate was estimated at 2.3%. Kwara State where Offa is located has a population of 2.37 million from the 2006 census (PHC, 2006).
Study design and periods
This project work was a cross sectional descriptive study in which subjects were hospital wastewater samples collected from the study site between the periods of April to October, 2018 and November, 2018 to March, 2019.
Collections of hospital wastewater samples
Two sets of twenty-one (21) hospital wastewater samples were collected from Offa Local Government Area of Kwara State, Nigeria. The first set of twenty-one samples (wet season samples) were collected between the months of April – October, 2018, while the second set of twenty-one samples (dry season samples) were collected between the months of November, 2018 – March, 2019. The samples were collected from hospital laboratory units into wide mounted sterile plastic containers with screw cap tops (universal bottles) corked tightly. The containers were labeled with date, time and sites of collection, and transported inside ice packs to Microbiology Laboratory, Obakekere, FUTA for culturing of bacteria.
Isolation and enumeration of bacterial colonies from hospital wastewater samples
Five-fold serial dilution was carried out on collected hospital wastewater samples. Aliquot (1 ml) of the diluents were pipetted into Petri-dishes, pour-plated with about 20 ml of molten nutrient agar at about 45°C and allowed to gel. The isolation of bacteria from wastewater was done according to methods of Cheesbrough (2010). The emerged colonies were counted using a colony counter and values were recorded after 24 h of incubation (Marshia et al., 2016).
Preparation of pure isolates of bacterial colonies from hospital wastewater samples
A distinct colony was taken and streaked with sterile wire loop on a freshly prepared solidified nutrient agar and incubated for 24 h at 37°C to get pure and distinct colonies. This was repeated several times until satisfactory pure isolates were obtained (Marshia et al., 2016).
Conventional identification of bacterial isolates in hospital wastewater samples
The identity of all isolates was determined using standard conventional methods as reported by Cheesbrough (2010). The bacterial isolates were cultured on nutrient agar and incubated at 37°C for 24 h and subsequently sub-cultured on to differential selective media namely Eosin Methylene blue agar and MacConkey agar. The bacterial isolates were tentatively identified by means of morphological characteristics, cellular and biochemical tests. Morphological characteristics were observed for each bacterial colony after 24 h of growth. The appearance of the colony of each isolate on the media was studied and the characteristics observed included cell shape, elevation, edge, optical characteristics, consistency colony surface and pigmentation. Biochemical tests carried out include; catalase, production of hydrogen sulphide (H2S), indole, urease, methyl red, oxidase, coagulase, motility, citrate utilization, methyl red, voges-proskauer, starch hydrolysis and sugar fermentation. The results were compared with Bergey’s Manual of Determinative Bacteriology (Fawole and Oso, 2007).
Molecular identification of bacterial isolates
Extraction of (deoxyribonucleic acid) DNA using cetyl trimethyl ammonium bromide (CTAB) method
Deoxyribonucleic acid (DNA) was extracted from hospital wastewater isolates by a standard CTAB genomic DNA isolation method as follows: 1 ml of 24 h broth culture was transferred into 1.5 ml Eppendorf tube and spun at 14,000 rpm for 30 min (to harvest the cell). 400 µl of a pre-warmed CTAB buffer (at 60°C) containing proteinase k and β-mercapto ethanol was added. Thereafter, 75 µl of 10% SDS (sodium deodycylsulphate) was added and heated in water bath at 65°C for 30 min. Five hundred micro liter (500 µl) chloroform was added, mixed for 15 min (to purify the DNA) and spun at 10,000 rpm for 10 min. The supernatant was collected in Eppendorf tube to which 500 µl isopropanol and 1 µl (100 mg/ml) RNase were added and incubated for 30 min at 37°C. The resultant mixture was kept at -20 for 24 h and spun at 10,000 rpm for 10 min. The supernatant was gently decanted and the pellet washed with 200 µl of 70% ethanol, gently mixed and spun at 10,000 rpm for 5 min. The extracted DNA was air-dried for 30 min to 1 h (to eliminate all traces of alcohol) and finally re-suspended in 200 µl of sterile distilled water (Doyle and Doyle, 1990).
Quantification of extracted deoxyribonucleic acid (DNA)
Quantification of DNA concentration and purity of the samples were measured using Nano-Drop® 2000 spectrophotometer. The ratio of 260/280 absorbance was used to assess the purity of DNA with ratios ~1.8 being accepted as pure (Akinyemi and Oyelakin, 2014).
Polymerase chain reaction (PCR) analysis of 16S
Polymerase chain reaction (PCR) analysis was run with a universal primer called 16S. The PCR mix comprises 1 µl of 10x buffer, 0.4 µl of 50 mM MgCl2, 0.5 µl of 2.5 mMdNTPs, 0.05 µl of 5 units/µlTaq with 2 µl of template DNA and 6.05 µl of distilled water to make-up 10 µl reaction mix. The PCR profile used is initial denaturation temperature of 94°C for 3 min, followed by 30 cycles of 94°C for 60 s, 56°C for 60 s, and 72°C for 120 s. The final extension temperature was 72°C for 5 min and the 10°C on hold for few hours (Akinyemi and Oyelakin, 2014).
Standardization of inoculums for antibiotic sensitivity test (0.5 McFarland standard)
About 0.1 ml of 1% barium chloride was added to 9.9 ml of 1% surphuric acid which was later reconstituted into 10 ml of sterile distilled water to make 0.5 ml McFarland standard solution. The broth culture of 24 h test organism was then compared in terms of turbidity to 0.5% McFarland. A loopful of the standardized culture was used for antibiotic sensitivity assay (Andrew, 2006; Paramedics World, 2018).
Antibiotics sensitivity test
The antibiotic sensitivity of the bacterial species isolated was performed on Mueller-Hinton agar (MHA) (Merck) plates by disk diffusion method as described by the National Committee for Clinical Laboratory Standard Institute (CLSI, 2017). A 0.1 ml of each bacterial isolate was seeded into each of the Petri dishes containing Mueller-Hinton agar and allowed to stand for 30 min to enable the inoculated organisms to pre-diffuse. The commercially available discs containing the following antibiotics: ceftazidine (CAZ, 30 µg), cefuroxime (CRX, 30 µg), gentamicin (GEN, 10 µg), ceftriaxone (CTR, 30 µg), erythromycin (ERY, 5 µg) cloxacillin (CXC, 5 µg), ofloxacin (OFL, 5 µg), augmentin (AUG, 30 µg), cefixime (CXM, 5 µg), nitrofrantion (NIT, 300 µg) and ciprofloxacin (CPR, 5 µg) (Liverpool L9 7AR, UK) were aseptically placed on the surfaces of the sensitivity agar plates with a sterile forceps and incubated at 37°C overnight. Zones of inhibition after incubation were observed and the interpretation was made using susceptibility breakpoints of CLSI (2017). The diameters of the zone of inhibition around the disc were measured to the nearest millimeter using a metal caliper and the isolates were classified as sensitive, intermediate and resistant.
Data quality assurance
Sample collection, handling, transportation as well as microbiological analysis and interpretation of results were carried out following standard operating procedures (SOPs). Prior to the actual work reagents, media and antimicrobial disks were checked for expiry date, damage and storage problems. Laboratory equipment were properly cleaned and sterilized before use. Media preparation was made based on the respective manufacturer’s directives. Five percent of media per batch/prepared was incubated overnight for sterility check.
Data analysis
Data obtained were analyzed using analysis of variance (ANOVA) and mean separated using Duncan’s Mean Multiple Range Test (IBM-SPSS) 20 version). Differences were considered significant at p < 0.05.
RESULTS
Mean count of bacteria isolated from hospital wastewaters
Generally, there were significant differences (p<0.05) in means count of bacteria in all wastewaters samples. Samples from Site C had the highest bacterial count (225 ±67.27 x 105 cfu per ml) during the dry season. On the other hand, during the wet season, Site A had the highest (150 ±43.59 x 105 cfu per ml) and Site K had the lowest bacterial count (7 ±4.00 x 105 cfu per ml) during the wet season (Table 1).
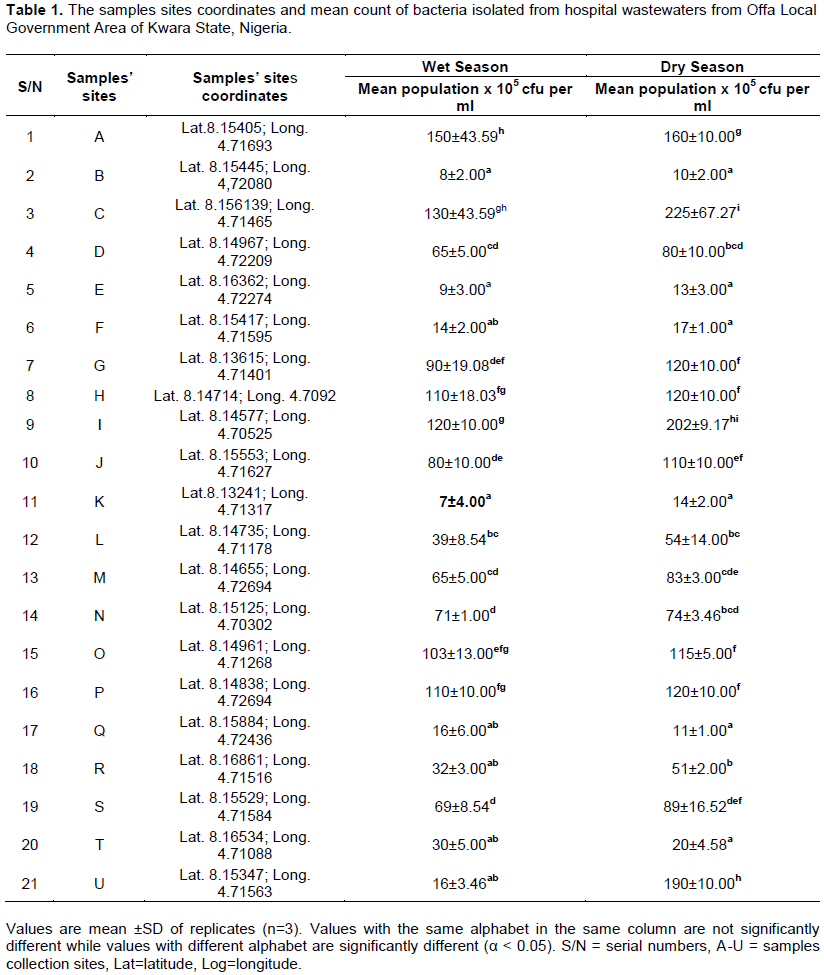
Bacteria isolated from hospital wastewaters
Alcaligenes faecalis was found present almost in the entire forty-two (42) sample sites (both during the dry and wet season periods), except in sites O and P (during the dry season), and sites J, O, Q and R (during the wet season). Staphylococcus saprophyticus were found present only in sites O and P (during the dry season), and also in sites O, Q and R (during the wet season). However, Alcaligenes aquatilis was found alone in site J during wet season (Table 2).
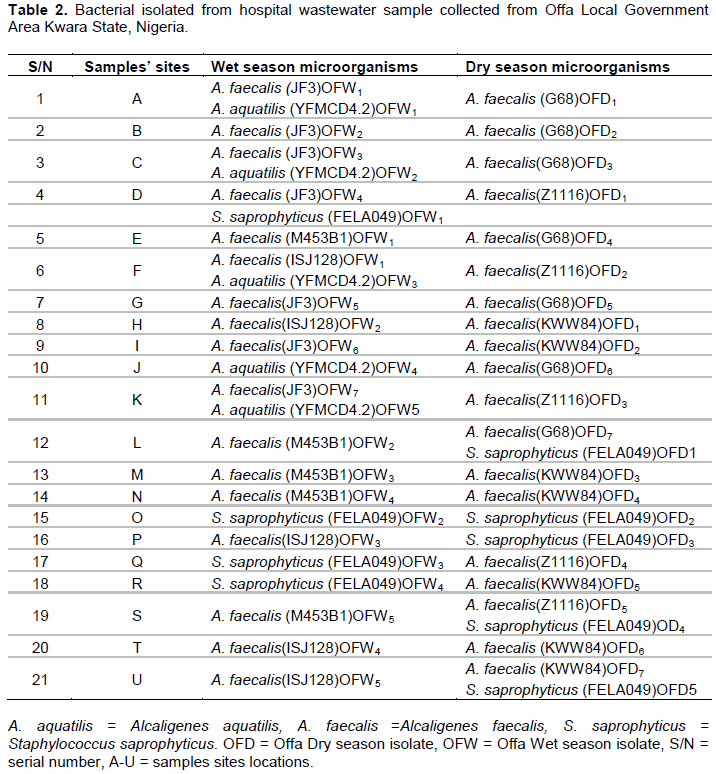
Prevalence/Percentage of bacterial isolates
A. faecalis made up to 19(79.2%) of the total bacteria isolated during the dry season and 17(65.4%) during the wet season while A. faecalis strain JF3 is more prevalent with percentage occurrence 26.9% during the wet season and strain G68 and KWW84 with percentage occurrence of 29.2 during the dry season (Table 3).
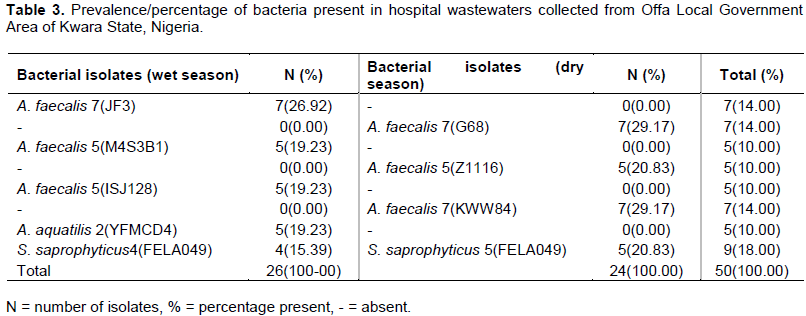
Antibiotics resistance pattern of bacterial isolates from hospital wastewater
The in-vitro antibiotic resistance pattern of the bacteria isolated from hospital wastewater indicated that all bacteria isolated were susceptible to ofloxacin during the dry and wet season periods, but were resistant to ceftazidine, cefuroxime, erythromycin, cloxacillin, augmentin and cefixime (Tables 4 and 5).
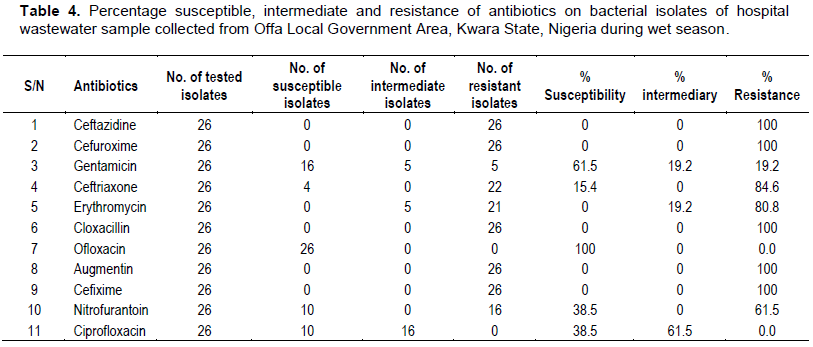
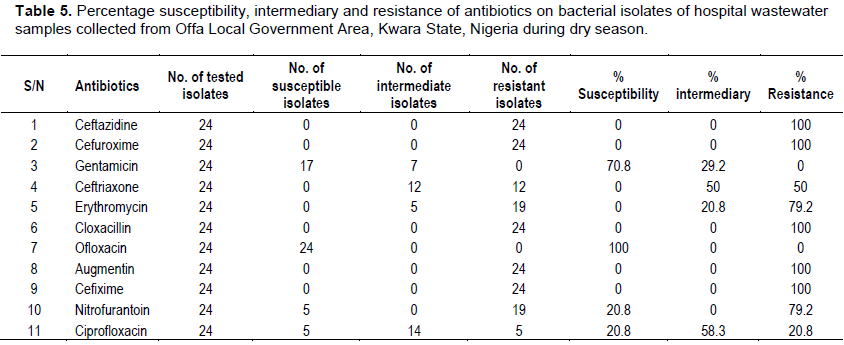
Antibiotics sensitivity profile of bacterial isolates from hospital wastewater
All bacterial isolates of hospital wastewater collected both during the wet and dry season periods were found to be 100% resistant to ceftazidine, cefuroxime, cloxacillin, augmentin and cefixime. The isolates were equally found to be 100% sensitive to ofloxacin. A. faecalis strain JF3, A. faecalis strain ISJI28 and S. saprophyticus strain FELA049 were found to be 100% sensitive to gentamicin, while only A. faecalis strain M4S3B1 was 100% resistant to gentamicin. Also, among the isolates, only A. faecalis strain M4S3B1 and Z1116 were 100% sensitive to nitrofurantoin and ciprofloxacin (Tables 6 and 7).
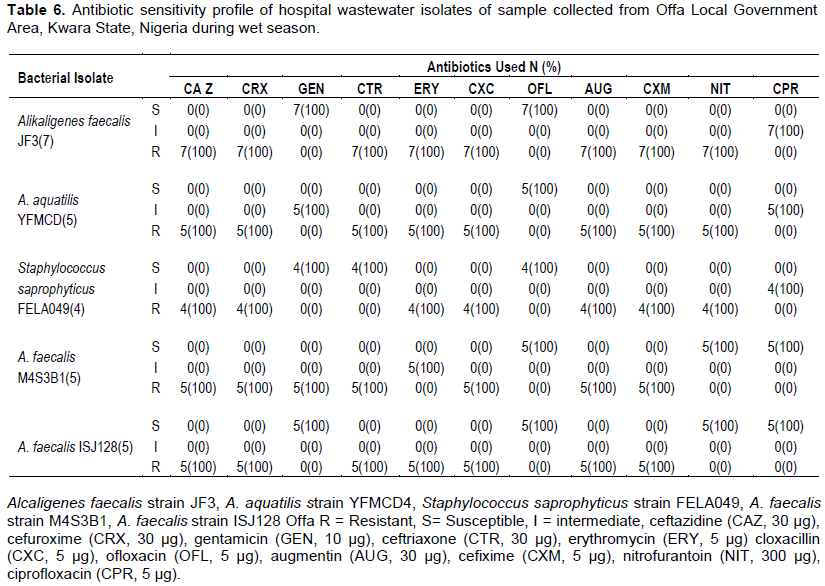
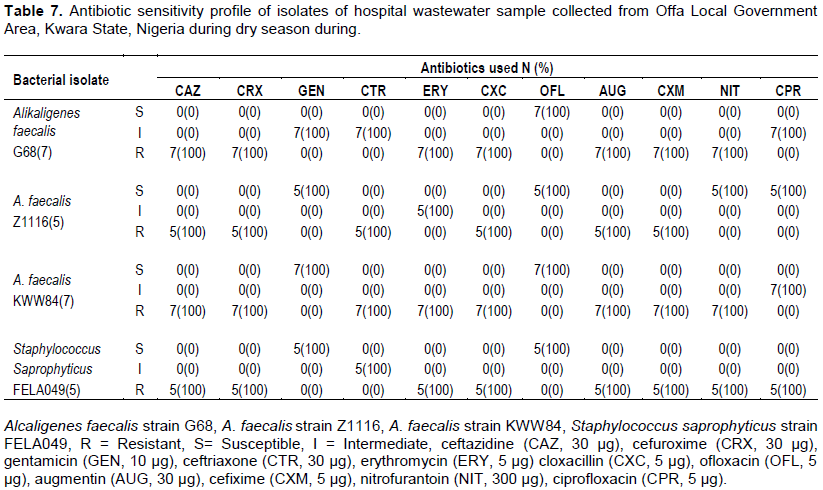
Total antibiotics sensitivity profile of bacterial isolates of hospital wastewater samples
The total resistance of bacterial isolates from hospital wastewater collected during the wet season was higher for ceftazidine, cefuroxime, cloxacillin, augmentin and cefixime 26/26 (100%) followed by ceftriaxone 22/26 (84.6%), erythromycin 21/26 (80.8%) and nitrofurantoin 16/26 (61.4%). However, relatively lower resistances were observed among bacterial isolates to gentamycin 5/26 (19.2%), ofloxacin 0/26 (0%) and ciprofloxacin 0/26 (0%).
Similarly, higher resistance was recorded during dry season for ceftazidine, cefuroxime, cloxacillin, augmentin and cefixime 24/24 (100%) followed by erythromycin 19/24 (79.2%), nitrofurantoin 19/24 (79.2%) and ceftriaxone 12/24 (50%). However, relatively lower resistance was observed among bacterial isolates to ciprofloxacin 5/24 (20.8%) (Figures 1 and 2).
DISCUSSION
The values of bacterial plate counts recorded during the dry and wet season periods in this research exceeded the permissible limit of Environment Protection Agency, EPA (2002) and Health Protection Agency, HPA (2005) (<1000 cfu/ml) and also failed to fulfill the requirements of the revised guidelines on the quality of treated wastewater used in agriculture, in public parks (<5 × 103cfu/100 ml) (Carr et al., 2004). High density of bacteria recorded both during wet and dry season periods, was an indication of environmental pollution due to human activities. This finding agrees with results recorded by Tsegahun et al. (2017) on wastewater at Ayder Referral Hospital, Mekelle North Ethiopia. Also, there were significant differences (p<0.05) in the means count of bacteria in all wastewater samples analyzed.
The variations observed in the values of the mean bacterial populations among hospitals in Offa Local Government Area may be due to variation in the rate of people’s patronage at different hospitals which is dependent on location, accessibility, health care facility and personnel available. Also, it was observed that higher density of microbial population was obtained during dry season than wet season. Therefore, preference of specific microorganisms to specific temperature ranges for growth and activity can have impacts on the composition of the microbial community (Fierer and Schimel, 2003; Singh et al., 2010).
The bacterial isolates found present in this study were different from that obtained by Tsegahun et al. (2017) on wastewater from Ayder Referral Hospital, Mekelle North Ethiopia, where Klebsiella spp, Pseudomonas aeruginosa, S. aureus, E. coli and Salmonella spp. were detected. The findings in the study of Tsegahun et al. (2017) also disagrees with the observation of Fekadu et al. (2015) who reported presence of Salmonella spp., Shigella spp., E. coli and S. aureus from effluent collected from Hawassa University Referral Hospital, Ethiopia. Also, isolates in this study were different from the study in India by Chitnis et al. (2000) that showed large numbers of enteric-bacteria S. aureus and P. aeruginosa. It is also dissimilar to work of Danchaivijitr et al. (2005) and Salem et al. (2011) who claimed availability of pathogenic bacteria like Vibrio spp. and Salmonella spp. in Thailand and Tunisia hospital effluents respectively. However, the findings of this study agreed with the work of Tsegahun et al. (2017) that confirmed the presence of S. aureus and CoNS (coagulase negative Staphylococcus) in treated hospital wastewater collected from Ayder Referral Hospital, Mekelle North, Ethiopia. The absence of some pathogenic bacteria in the hospital wastewater analyzed may be due to variation in geographical and climatically condition as shifting of microbial community occurs in favour of the species which are better adapted to higher temperatures and have accelerated rates of growth (Castro et al., 2010; Singh et al., 2010; Fierer and Schimel, 2003). More so, inter-specific competition among microorganisms may cause shift in microbial community, such that microorganisms that compete favorably among the mixed community due to several factors such as population density, inhibitory metabolites, and so on will be prevailing. The highest prevalence of A. faecalis may be due to the fact that it is highly associated with urinary tract infection (UTI) which is common in hospital environment, and production of toxic exudates might also favour survival of Staphylococcus spp.
The resistance of all the bacterial isolates to ceftazidine, cefuroxime, cloxacillin, augmentin and cefixime was similar to finding of Katouli et al. (2012) where isolates showed simultaneous resistance for ampicillin with clavulanic acid, cotrimoxazole, tetracycline, first, second and third generation cephalosporins in the final effluent of wastewater treatment plant in India. Study in Alexandria, Egypt also showed the presence of antibiotic resistant extended spectrum beta-lactamase (ESBL) producing bacteria at the end of wastewater purification process (Amine, 2013), posing a risk of its spread to the environment and subsequent human and animal exposure. Overall resistance of bacteria isolated during the wet season for ceftriaxone, erythromycin, nitrofurantoin and gentamycin were found to be 84.6, 80.8, 61.4 and 19.2% respectively. Similarly, all bacterial isolates from hospital wastewater collected during the dry season were found to be 100% resistant to ceftazidine, cefuroxime, cloxacillin, augmentin and cefixime, and were equally found to be 100% sensitive to ofloxacin. Similar observation was reported by Iweriebor et al. (2015) from Alice, Eastern Cape province of South Africa and European countries. Also, Servais and Passerat (2009) confirmed higher rate of resistance in bacterial isolates from final effluent of wastewater treatment plant. The resistance of microbes to these antibiotics may be due to abuse of these antibiotics by their users or their failure to adhere to instruction given by the physicians (Davey et al., 2002). More so, presence of high percentage of drug resistant isolates from hospital wastewater suggests that, hospital wastewater could have contributed massively to the resistances observed among the isolates from the final effluent. These can be due to the fact that, hospital wastewater contains a diverse group of pathogenic commensals and environmental bacteria. This characteristic composition makes sewage particularly suitable ecological niche for the growth and spread of antibiotic resistance due to selection pressure and horizontal gene transfer (Davies and Davies, 2010; Periasamy and Sundaram, 2013; Cantona et al., 2013).
CONCLUSION AND RECOMMENDATIONS
A. faecalis was the most predominant among the isolates from hospital wastewater samples analysed followed by S. saprophyticus, which exceeded the WHO, HPA, EPA and FAO standard permissible levels. The high prevalence of drug resistant isolates from hospital wastewater samples analyzed suggests their persistence in the hospital environment, and their ability to spread antibiotic resistance due to selection pressure and horizontal gene transfer. Therefore, patients are advised to adhere strictly to the directives of the physician in administration of drugs so as to reduce the cases of antibiotics resistance. Also, adequate liquid waste treatment system should be developed to disinfect pathogens in hospital wastewater effluent before discharging into municipal water supply, so as to prevent diseases associated with hospital wastewater effluent microbes.
CONFLICT OF INTERESTS
The authors have not declared any conflict of interests.
REFERENCES
|
Abdel-Rouf N, Al-Homaidan AA, Ibraheem IBM (2012). Microalgae and wastewater treatment. Saudi, Journal of Biological Science 19(3):257- 275. |
|
|
Akinyemi AA, Oyelakin OO (2014). Molecular characteristics of bacteria isolated from farm-raised catfish Clarias grariepinus. British Microbiology Research Journal 4(12):1345-1352. |
|
|
Amine AEK (2013). Extended spectrum Beta-lactamase producing bacteria in wastewater, Alexandria, Egypt. International Journal of Bioscience, Biochemistry Bioinformatics 3(6):605-608. |
|
|
Andrew JM (2006). BSAC Standardized disc susceptibility testing methods. Journal of Antimicrobial Chemotherapy 58(3):511-529. |
|
|
Anitha J, Jayraaj IA (2012). Isolation and identification of bacteria from biomedical waste (BMW). International Journal of Pharmacy and Pharmaceutical Sciences 4(5):0975-1491. |
|
|
Aurelien BDH, Sylvie B, Alain D, Jerome G, Yves P (2013). Ecotoxicological risk assessment linked to the discharge by hospitals of bio-accumulative pharmaceuticals into aquatic media: The case of mitotane, Chemosphere 93(10):2365-2372. |
|
|
Beyene H, Redaie G (2011). Assessment of waste stabilization ponds for the ttreatment of hospital wastewater: The case of Hawassa University Referral Hospital. World Applied Science Journal 15(1):142-150. |
|
|
Cantona R, Horcajadad JP, Oliverb A, Garbajosaa PR, Vilab J (2013). Inappropriate use of antibiotics in hospitals: The complex relationship between antibiotic use and antimicrobial resistance. Enfermedades Infecciosasy Microbiologia Clinica 31:3-11. |
|
|
Carr RM, Blumenthal UJ, Mara DD (2004). Guidelines for the safe use of wastewater in agriculture: Revisiting WHO Guidelines. Water Science and Technology 50(2):31-38. |
|
|
Castro HF, Classen AT, Austin EE, Norby RJ, Schadt CW (2010). Soil microbial community responses to multiple experimental climate change drivers. Applied and Environmental Microbiology 76(40):999-1007. |
|
|
Cheesbrough M (2010). District laboratory practice in tropical countries, 3rd edition, Cambridge, University Press, United Kingdom pp. 50-176. |
|
|
Chitnis V, Chitnis, D, Patil S, Kant R (2000). Hospital effluent: A source of multiple drug-resistant bacteria. Current Science 79:989-991. |
|
|
Clinical Laboratory Standards Institute (CLSI) (2017). Performance standards for antimicrobial susceptibility testing; Twenty-fourth informational supplement. CLSI document M100-S24 Wayne, PA: ISBN 1-56238-898-3. 34(1):50-98. |
|
|
Danchaivijitr S, Wongchanapai W, Assanasen S, Jintanothaitavorn D (2005). Microbial and heavy metal contamination of treated hospital wastewater in Thailand. Journal of the Medical Association of Thailand 88:59-64. |
|
|
Davey P, Pagliar C, Haves A (2002). The patient's role in the spread and control of bacterial resistance to antibiotics. Clinical Microbiology and Infection 8(2):43-68. |
|
|
Davies J, Davies D (2010). Origins and Evolution of Antibiotic Resistance. Microbiology and Molecular Biology Reviews 74(3):417-433. |
|
|
Diwan V, Tamhankar AJ, Khandal RK, Shanta S, Aggarwal M, Marothi Y, Rama VL, Sundblad-Tonderski K, Stalsby-Lundborg C (2010). Antibiotics and antibiotic resistant bacteria in waters associated with a hospital in Ujjain, India. BMC Public Health 10(1):414-422. |
|
|
Doyle JJ, Doyle JL (1990). Isolation of plant DNA from fresh tissue. Focus 12:13-15. |
|
|
Environment Protection Agency (EPA) (2002). US Environment Protection Agency, Safe Drinking Water Act Amendments. |
|
|
Fawole MO, Oso BA (2007). Laboratory manual of microbiology. 5thedition. Spectrum Books limited, Ibadan, Nigeria pp. 22-23. |
|
|
Fekadu S, Merid Y, Beyene H, Teshome W, Gebre-Selassie S (2015). Assessment of antibiotic and disinfectant resistant bacteria in hospital wastewater. South Ethiopia. Journal of Infection in Developing Countries 9(2):149-156. |
|
|
Ferrando-Climent L, Rodriguez-Mozaz S, Barcelo D (2014). Incidence of anticancer drugs in an aquatic urban system: from hospital effluents through urban wastewater to natural environment. Environmental Pollution 193:216-223. |
|
|
Fierer N, Schimel JPA (2003). Proposed mechanism for the pulse in carbon dioxide production commonly observed following the rapid rewetting of a dry soil. Soil Science Society of America Journal 67(3):798-805. |
|
|
Gautam AK, Kumar S, Sabumon PC (2007). Preliminary study of physico-chemical treatment options for hospital wastewater, Figure 2: Heavy metal concentrations in different sampling locations (a) Mn (b) Pb43. Journal of Environmental Management 83(3):298-306. |
|
|
Health Protection Agency (HPA) (2005). The microbiological examination of water samples, National Standard Method QSOP 57, Issue 2. |
|
|
Iweriebor BC, Gaqavu S, Chikwelu OL, Nwodo UU, Okoh A (2015). Antibiotic susceptibilities of Enterococcus species isolated from hospital and domestic wastewater effluents in Alice, Eastern Cape Province of South Africa. International Journal of Environmental Research and Public Health 12(4):4231-4246. |
|
|
Katouli M, Thompson JM, Gundogdu A, Stratton HM (2012). Antibiotic resistant bacteria in hospital wastewaters and sewage treatment plant. Science Forum and Stakeholder Engagement: Building Linkages, Collaboration and Science Quality13:225-229. |
|
|
Keen PL, Patrick DM (2013). Tracking Change: a look at the ecological footprint of antibiotics and antimicrobial resistance. Antibiotics Review 2:191-205. |
|
|
Marshia P, Mir Rowshan A, Mostafizer R, Fakhruzzamania (2016). Isolation and identification of bacteria with determination of bacterial loads from different brands of butter and cheese. Asian-Australasian Journal of Bioscience and Biotechnology 1(3):504-513. |
|
|
Offa Descendants Union of North America (ODUNA) (2020). History of Offa. |
|
|
Oteo J, Campos J, Baquero F (2001). Antibiotic resistance in 1962 invasive isolates of Escherichia coli in 27 Spanish Hospitals participating in the European antimicrobial resistance Surveillance System. Journal of Antimicrobial and Chemotherapy 50(6):945-952. |
|
|
Pandey A, Afsheen AF, Kumar TSK (2011). Isolation and characterization of multidrug resistance cultures from wastewater. Journal of Pharmaceutical Biomedical Science 13(7):1-7. |
|
|
Paramedics World (2018). Preparation of McFarland standard for antibiotic susceptibility test (AST) in laboratory. GoogleChrome.pramedicsworld.com Retrieved on 10th January, 2021. |
|
|
Periasamy D, Sundaram A (2013). A novel approach for pathogen reduction in wastewater treatment. Journal of Environmental Health Science and Engineering 11(1):12-21. |
|
|
Population and Housing Census (PHC) (2006). Federal Republic of Nigeria - IHSN survey catalog, 2006 population and housing census, priority table III, population distribution by sex, state LGA and senatorial district P 38. |
|
|
Salem IB, Ouardani I Hassine, Maouni M (2011). Bacteriological and physico-chemical assessment of wastewater in different region of Tunisia: Impact on Human Health. BMC Research Notes 4(1):144-155. |
|
|
Servais P, Passerat J (2009). Antimicrobial resistance of faecal bacteria in waters of Seine River Watershed (France). Science of the Total Environment 408(2):365-372. |
|
|
Singh BK, Bardgett RD, Smith P, Reay DS (2010). Microorganisms and climate change: terrestrial feedbacks and mitigation options. Nature Review Microbiology 8(11):779-790. |
|
|
Stalder T, Barraud O, Jove T, Casellas M, Gaschet M, Dagot C, Ploy MC (2014). Quantitative and qualitative impact of hospital effluent on dissemination of the integron pool. The ISME journal 8(4):768-777. |
|
|
Tsegahun A, Letemichael N, Amlsha K, Yemane W (2017). Antibiotic resistant bacteria from treated and untreated hospital wastewater at Ayder Referral Hospital, Mekelle, North Ethiopia. Advances in Microbiology 7(12):871-886. |
|
|
Verlicchi P, Galletti A, Petrovic M, Barcelo D (2010). Hospital effluents as a source of emerging pollutants: an overview of micro-pollutants and sustainable treatment options. Journal of Hydrology 389(3-4):416-428. |
|
|
Wikipedia (2020). Offa - Kwara State. |
|
|
Wilde ML, Mahmoud WMM, Kümmerer K, Martins AF (2013). Oxidation-coagulation of β-blockers by K2FeVIO4 in hospital wastewater: Assessment of degradation products and biodegradability, Science of the Total Environment 452(453):137-147. |
|
Copyright © 2024 Author(s) retain the copyright of this article.
This article is published under the terms of the Creative Commons Attribution License 4.0