Photosynthetic bacteria (PSB) are a group of phototrophic anoxygenic prokaryotic organisms that are distributed widely in nature. PSB contains rich pigments such as bacteriochlorophyll and carotenoids (Oudin et al., 2007). The remarkable physiological functions of carotenoids include the following: they act as antioxidants to quench singlet oxygen and capture oxygen free radicals, are nutritional supplements, food pigments and feed colorants in industry (Hirschberg, 2001). Most commercial carotenoids are obtained by chemical synthesis, but the growing demand for natural additives has spurred several metabolic engineering approaches to increase carotenoids production in plants and microorganisms (Carrera et al., 2007). About 80 different carotenoids are synthesized by photosynthetic bacteria. All the carotenoids in photosynthetic bacteria are bound to light-harvesting or reaction center complexes (Zhou et al., 2015). As early as 1970s, the chemical structure of carotenoids was elucidated, and their biosynthetic pathways were researched demonstratively and clearly (Viveiros et al., 2000). Structurally carotenoids are tetraterpenes derived from the 5-carbon unit isopentenyl diphosphate (IPP) and its isomer dimethylallyl diphosphate (Eisenreich et al., 2001). There are two pathways for the synthesis of these carotenoids: the mevalonic acid (MVA) pathway and the methylerythritol 4-phosphate (MEP) pathway. The MEP pathway uses glyceraldehy 3-phosphate and pyruvate as initial substrates (Sandmann, 1994). In recent years, people have been devoted to the cloning and analysis of genes related to carotenoid biosynthesis pathway in microorganisms and plants; in particular, huge progress has been made in modifying plant carotenoid biosynthesis pathway through genetic engineering. Although some intermediates, mechanisms, enzymes and genes of this pathway have been identified, many details of this widely distributed pathway have not been identified (Viveiros et al., 2000).
The synthesis of carotenoids in PSB is a highly coordinated biological process. Its metabolic pathways are complex, and can easily be interrupted by other metabolites. The external environmental factors such as the intensity of light and temperature also greatly affect the accumulation of pigments (Guan et al., 2015). Therefore, the study of bacterial carotenoid biosynthetic pathways is very important.
Metabonomics provides a comprehensive description of all metabolite levels in organisms and has been increasingly used to characterize microbial characteristics, such as cell stress response, high yield mechanisms, and microbial complex interactions. The differences in the fermentation processes of microorganisms at the metabolic level can provide unique insights into the study of metabolic pathways (Okuda et al., 2008). Large differences in the type and quantity of metabolites are often key limiting factors for microbial fermentation. However, there are very few reports on carotenoids synthesized by bacteria around the world. In this study, metabolomics based on LC-MS technology was used to detect the metabolites of the fermentation of PSB-W under different fermentation times and different light intensity treatments. Through comparative metabolomics analysis, the key metabolites for carotenoid synthesis were found, and the carotenoid synthesis mechanism was initially established in combination with the KEGG database, providing a basis for further research on carotenoid metabolism engineering.
Bacterial strain and growth conditions
A photosynthetic bacterium P. acetatigenes PSB-W was isolated from a water purifying agent stored in the Microbiology Laboratory of Shanxi Normal University. The PSB-W cells were cultured on a basic medium consisting of (in g/L) NH4Cl (1.5), K2HPO (0.5), NaHCO3 (2.0) and yeast extract (3.5). The initial pH value was adjusted to 6.8-7.2. Cells were activated under the condition of incubation at 30°C and 60-W tungsten lamp illumination of 1500 Lux for 4 days.
Sample preparation
Duration of fermentation
The activated strain from above was transferred to another basic medium for fermentation to accomplish a concentration of 10% inoculum; it was cultured in light-anaerobic conditions at different times. Fermentation broth was taken at the 48, 60, 72, 96 h as the analysis and experimental objects. The light intensity was kept at 1500 Lux.
Light intensity
The activated strain was transferred to the fermentation medium (10% inoculum amount). It was placed in incubators with light intensities of 1000 Lux and 1500 Lux respectively for anaerobic culture for 4 days to obtain the fermentation liquid as samples. There were three replications at each light intensity.
Metabolites extraction
Samples were thawed at 4°C on ice and then extracted with extraction solvent (methanol: acetonitrile, 1:1, V:V), followed by vortexing for 30 s, ultrasound treatment for 5min, and incubated for 1h at -20℃ to precipitate proteins. These were centrifuged at 12000 rpm for 15 min at 4°C; then the supernatant was collected and the pellet was discarded. The supernatant was dried in a vacuum concentrator without heating, and solvent extraction (acetonitrile: water, 1:1, V:V) reconstitution was added. It was vortexed for 30 s and sonicated for 10 min, and centrifuged for 15 min at 12000 rpm. The supernatant was transferred for the UHPLC-QTOF-MS analysis.
LC-MS/MS analysis
LC-MS/MS analyses were performed using an UHPLC system (1290, Agilent Technologies) with a UPLC BEH Amide column (1.7 μm 2.1*100 mm, Waters) coupled to Triple TOF 6600 (Q-TOF, AB Sciex). The mobile phase consisted of 25 mM NH4Ac and 25 mM NH4OH in water (A) and acetonitrile (B) was carried with elution gradient delivered at 0.5 mL min-1. The injection volumes were 1 μL. The Triple TOF mass spectrometer was used for its ability to acquire MS/MS spectra on an information-dependent basis (IDA) during an LC/MS experiment. In this mode, the acquisition software was Analyst TF 1.7, AB Sciex, collision energy (CE) 30 V, ion source gas as 60 Psi, source temperature 600°C, Ion Spray Voltage Floating (ISVF) 5000 V or -4000 V in positive or negative modes respectively.
Data processing and statistical analysis
MS raw data (.wiff) files were converted to the mzXML format using ProteoWizard, and processed by R package XCMS (version 3.2). R package CAMERA was used for peak annotation after XCMS data processing. In-house MS2 database was applied in metabolite identification. Multivariate statistical analysis was performed with principal component analysis (PCA) to distinguish the samples according to metabolic similarity and with the Volcano Plot model to screen the main differential metabolites related to carotenoid metabolism pathway with respect to the classification. These were combined with KEGG database to classify the metabolism pathways of the carotenoids.
Metabolomics data analysis of fermentation broth at varying culture durations
Fermentation liquid composition analysis
Different groups of fermentation broth samples incubated for varying durations (0, 48, 60, 72, and 96 h) were analyzed by LC-MS. The metabolite composition of fermentation broth at different culture time is shown in Table 1.
In total, 26 putative metabolites were detected, mainly sugars, amino acids, and organic acids. But alkanes and ketones were detected in the control group (0 h). In the fermentation broth of 48 h group, L-glutamic acid, aspartic acid, alanine and other amino acids and organic acids were mainly detected. Amino acids are important for cell growth and protein synthesis in microorganisms. It is speculated that this stage is the delayed period of bacterial growth which synthesizes a large number of substances related to life activities for adapting to the environment (Luo et al., 2015). Among them, glutamic acid is the synthesis precursor of purine, pyrimidine and base, and participates in many metabolic pathways as the provider of amino group in organisms. Alanine is made from glutamate by reverse transamination and can be converted back to glutamate by alanine transaminase (Guan et al., 2015). Glutamate decreases at 96 h, which may also be one of the reasons for the decreased alanine. Midazole-4-keto-5 propionic acid is an intermediate product of histidine metabolism, which can synthesize D-ribose through the HMP pathway in the process of histidine biosynthesis. During the process of bacterial synthesis of carotenoids through this pathway acetyl-CoA is formed and then further forms IPP. Subsequently, the presence of d-ribose was also detected in the 60 h group samples (Luo et al., 2015). After 60 h of culture, other new additions to the fermentation broth are acetic acid and diisobutyladipate. Acetic acid is one of the main byproducts of lactic acid fermentation. When the fermentation time increased to 72 h, secondary metabolites such as phenols and alcohols were detected in the fermentation broth, while the content of amino acids decreased. It is speculated that the bacteria enter a stable period and the demand for amino acids is reduced for the synthesis of bacteria. When the fermentation time reached 96 h the
variety of metabolites detected were significantly reduced, and complex chemical components of cyclic hydrocarbons were detected. This indicates that with the extension of the culture time, the consumption of nutrients in the fermentation medium increased, and the growth force was gradually weakened (Wilson et al., 2005).
In the early stage of bacterial fermentation, the synthesis of components related to self-growth is to adapt to the environment for primary metabolism. After entering the rapid growth stage, a large number of bacteria propagated and began to synthesize carotenoids, resulting in the rapid increase of pigment content. In the later stage, due to the depletion of nutrients in the culture medium, the accumulation of acids, alcohols and other components that are not conducive to the growth of bacteria in the fermentation broth slowed down the metabolism (Santos and Meireles, 2010). This result is consistent with the phenomenon that occurs during the fermentation process resulting in the change of fermentation liquid from light red to red until dark red. Some components may be produced in the metabolic activity and then be decomposed and utilized to participate in the whole life process of bacteria.
Principal component analysis
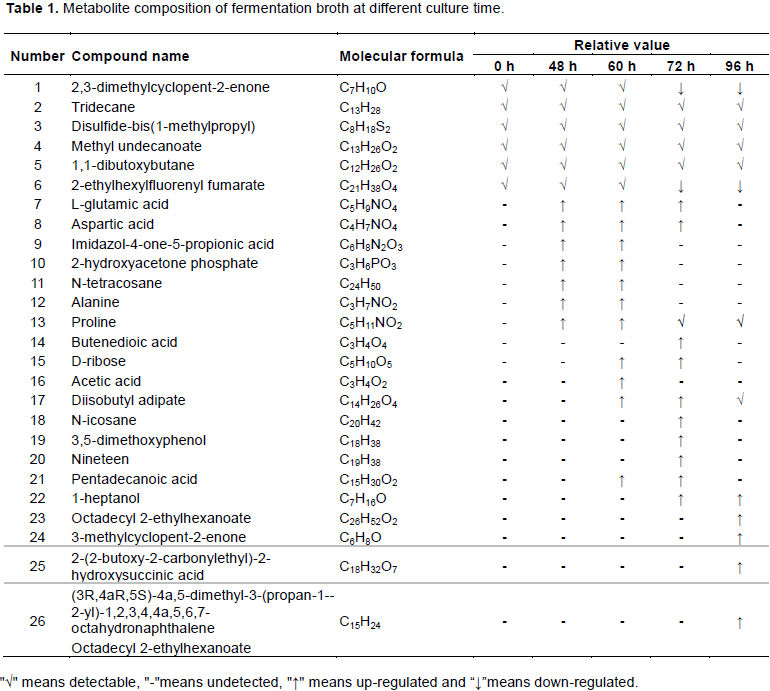
To establish the differences in the metabolites of the photosynthetic bacterial fermentation broth, metabolomics analysis was carried out. The data were processed with MZmine 2 software; multivariate statistical analysis was performed with PCA to discriminate the fermentation process of the strain through score plot (Santos and Meireles, 2010). It can be seen from Figure 1 that the contribution rate of principal component 1 (PC1) is 37%, and the contribution rate of principal component 2 (PC2) is 16%. This shows that the two principal components can represent 53% of metabolite variable information. The samples fermented for 48 and 60 h came together in a heap, and are in one category. The 72 and 96 h samples were grouped into a second category that is far apart from the first. It shows that the fermentation time has a great influence on the metabolites, and the changes of the metabolites have changed in stages with the change of time. The samples of the same group are tightly clustered together, indicating that the differences between the samples are small.
Metabonomics data analysis of fermentation broth at different light intensities
Fermentation liquid composition analysis
To assess the effect of light intensity on the fermentation broth, metabolomics analysis was performed. The bacteria were cultured for 4 days at light intensities of 1000 Lux and 1500 Lux respectively, which are relatively suitable for the growth of photosynthetic bacteria. LC-MS detection on the fermentation broth was done. A total of 37 metabolites were detected and analyzed, including sugars, amino acids and organic acids (Table 2). Only 10 compounds were detected in the 1000 Lux group, significantly less than 1500 Lux group in which 37 compounds were detected. This indicates that photosynthetic bacteria are more sensitive to light (Okuda et al., 2008). At low light intensities, carotenoid and other pigments capture insufficient light energy and cause bacterial metabolism to slow down and the synthesis of substances related to growth and reproduction is reduced. Compared with 1000 Lux group, amino acid components detected in the fermentation broth of 1500 Lux group were significantly increased, including proline, l-proline, l-leucine, o-acetyl-l-serine, valine, amongst others. It has been reported that amino acids play an important role not only in cell signaling, but also in regulating the phosphorylation cascade of gene expression proteins. In addition, these amino acid precursors are mostly intermediate metabolites of the TCA cycle and EMP pathway, so the increase of these intermediates may also cause the growth of carotenoids content (Sun et al., 2010). Proline may protect the bacteria by stabilizing the spatial structure of the enzyme, and also has the structure and function of stabilizing collagen, maintaining cell osmotic pressure and increasing salt and drought resistance. L-leucine is catalyzed to form α-ketoisocaproic acid by transaminase, and then is decarboxylated to form acetyl-CoA to participate in the citric acid cycle. Leucine is more efficient than other amino acids, has faster transamination rate and the ability to completely oxidize to produce adenosine triphosphate (Luo et al., 2015). In the 1500 Lux group, intermediate metabolites of some purine nucleotides such as hypoxanthine, deoxyadenine nucleoside, adenine, and xanthine were detected. Some of them can participate in the tricarboxylic acid cycle as the substrate of the tricarboxylic acid cycle, and quickly provide energy through the tricarboxylic acid cycle oxidation; another part of the intermediate products can be used as allosteric effectors of related enzymes in the process of energy metabolism, which can affect other metabolic pathways by regulating enzyme activity. High content of mannitol was also detected in 1500 Lux. Mannitol is a kind of photosynthetic assimilate, which can generate mannitol-1-phosphate under the catalysis of mannitol kinase and mannitol-1-phosphate dehydrogenase, and then generate fructose-6-phosphate to enter the glycolysis cycle and participate in energy metabolism. The results showed that compared with the 1000 Lux treatment group, the metabolites in the fermentation broth cultured under the light intensity of 1500 Lux were significantly increased. The results showed that the effect of light intensity on photosynthetic bacteria was extremely large, and the increase of light intensity could promote the rapid synthesis of cell energy materials by bacteria. Many metabolites in fermentation broth participated in the synthesis of carotenoid precursor IPP (Luo et al., 2015).

Principal component analysis
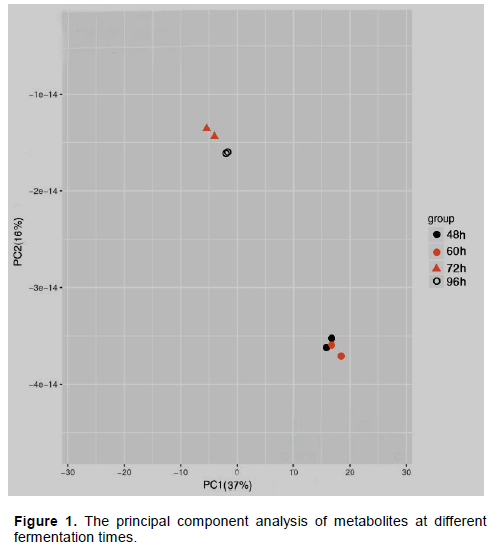
The metabolites were obtained from LC-MS detection of fermentation broth with different light intensity by PCA analysis. The results of the principal component score diagram are shown in Figure 1. In the score plot, three parallels for each sample clustered together, indicating that metabolomics was appropriate for monitoring the fermentation processes and interpreting the distinctive metabolic characteristics of the samples. The samples collected at different light intensity during fermentation can be clearly distinguished. This shows that the metabolites screened by score plot can be used as potential biomarkers (Steinmann and Ganzera, 2011). It can be seen from Figure 2 that the metabolites detected in the fermentation broth under two different light intensity treatments are extremely different. The samples of different light intensity groups are completely separated in the figure; instead, they are grouped together, indicating
that the metabolic differences between the treatment groups are significant.
Differential metabolite screening, functional annotation and enrichment analysis
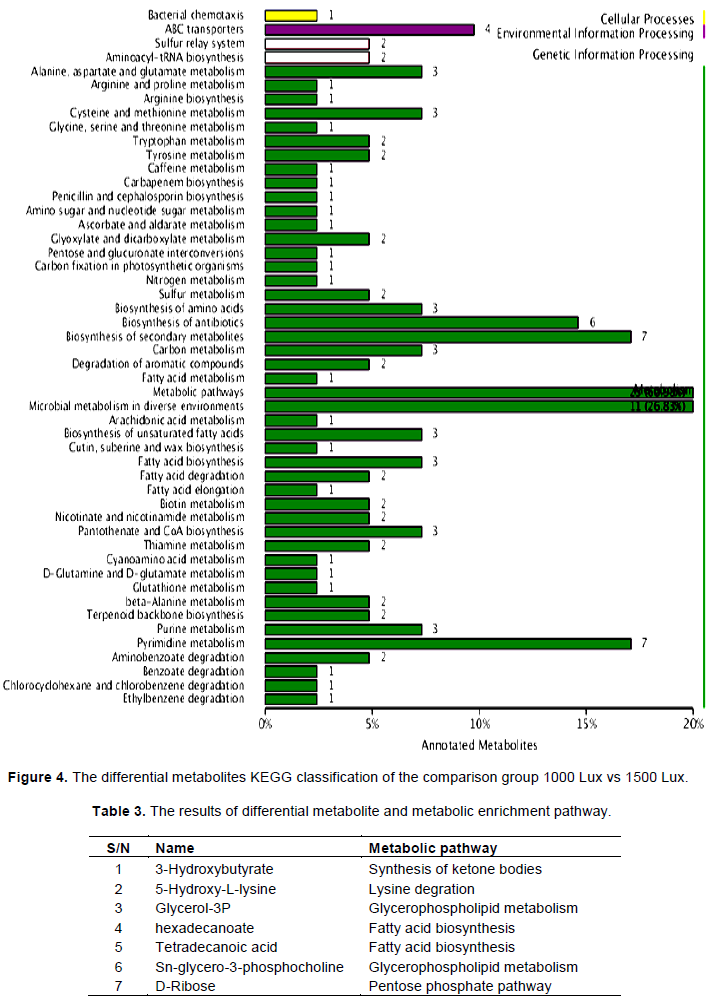
Differential metabolites were screened for each comparison group by combining the fold change and variable importance in project (VIP) values of the Volcano Plot model. The criteria for screening included the fold change value of ≥ 2 and the VIP value of ≥ 1. The screening results are shown in Figure 3. Concisely, there were 409 significant metabolites between 1000 Lux and 1500 Lux (273 down-regulated, 136 up-regulated).
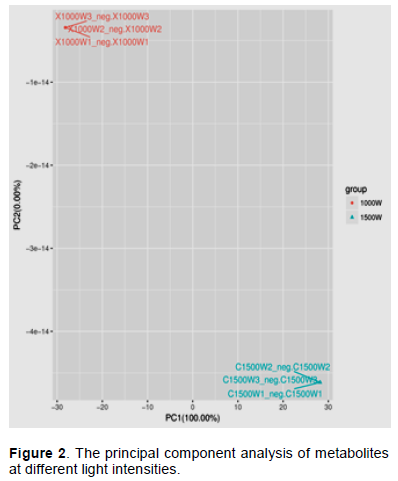
The differential metabolites were annotated by the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (Figure 4). The KEGG classification results indicated that the differential metabolites of comparison groups were involved in microbial metabolism in diverse environments, biosynthesis of secondary metabolites, pyrimidine metabolism, terpenoid backbone biosynthesis, carbon metabolism, biosynthesis of amino acids, pantothenate and CoA biosynthesis and other metabolic pathways. From among them, 7 differential metabolites related to the carotenoid synthesis enrichment pathway were screened (Table 3).
Pathway analysis
In order to unravel the relationships of these metabolites and their effects on the metabolic network, a possible synthetic pathway was identified for the selected differential metabolites in combination with the KEGG pathway database (Steinmann and Ganzera, 2011). The pathway model of carotenoids biosynthesis is depicted in Figure 5.
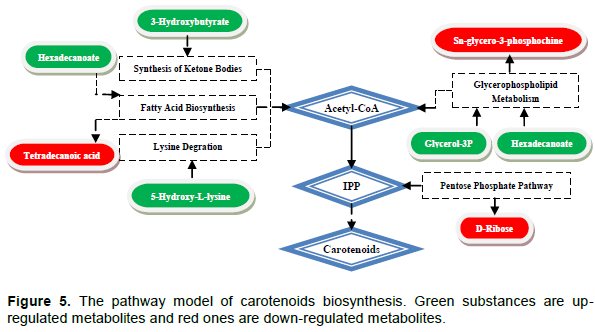
In Figure 5, 3-hydroxybutyrate is an up-regulated metabolite and an intermediate product in the synthesis of acetoacetate. It can be used as an intermediate product in the synthesis of acetoacetic acid and participate in the synthesis of IPP. 5-hydroxy-L-lysine can be degraded by lysine to form acetyl coenzyme A. D-ribose is related to the synthesis of the pentose phosphate pathway, bacteria produce glyceraldehyde 3-phosphate and phosphate ribose to convert to IPP through this pathway (Luo et al., 2015). It also produces a variety of important carbon-containing compounds (four-carbon, five-carbon) and multiple invertases. Sn-glycer-3-phosphocholine is a precursor substance that forms glycerol 3-phosphate and Glycerin-3-phosphate is the premise of glycerin formation. They are related to the pathway of glycerophosphatidic acid metabolism, which forms acetyl coenzyme A involved in pigment biosynthesis. Glycerol can regulate intracellular osmotic pressure as an activator of high osmotic pressure glycerol pathway. At the same time, glycerin-3-phosphate also comes from the EMP pathway, and the enhancement of the EMP pathway will also affect the levels of these two metabolites (Steinmann and Ganzera, 2011). In addition, 8 pyruvate molecules, 8 glyceraldehyde 3-phosphate molecules, 8 CTP molecules, 8 ATP molecules, and 16 NADPH molecules can synthesize a single molecule of lycopene. Tetradecanoic acid and hexadecanoate, these fatty acids undergo significant changes in 1500 Lux. This indicates that the biosynthesis of fatty acids is susceptible to environmental influences. In bacteria, the fatty acids in the peroxisome undergo b-oxidation; the resulting acetyl-CoA is converted into acetylcarnitine, and then enter the mitochondria for tricarboxylic acid (TCA) metabolism. The up-regulation of the intermediate products of the TCA cycle increased the metabolic flow of carotenoid synthesis; It is speculated that the intermediate metabolites of TCA are the carbon skeleton necessary for carotenoid synthesis. As carotenoids are intracellular metabolites, the accumulation of carotenoids increases with the bacteria growth.
In the present study, the key metabolic regulation nodes of carotenoid synthesis in PSB-W were identified through comparative metabolomics analysis of fermentation broth subjected to different fermentation times and light intensities. As an omics technology with the widest scope of application, metabolomics is unbiased to different samples and provides a systematic depiction of microbes at the integrated level. Several techniques have been developed for metabolomics analysis, such as capillary electrophoresis–mass spectrometry (CE–MS), nuclear magnetic resonance (NMR), gas chromatography–mass spectrometry (GC–MS), and LC–MS. LC–MS is a powerful alternative detection tool for metabolomics with the advantages of high separation efficiency, a simple pretreatment process, and high resolution (Steinmann and Ganzera, 2011). The results showed that the technique was mature and the metabonomics method was a powerful way of potential biomarker discovery (Helen et al., 2008). In this paper, we focused on the changes of PSB metabolites under different culture time and light intensity. Through the statistical analysis of samples, it was found that the synthesis of metabolites was staged, and the change of growth conditions would affect the normal synthesis and decomposition of metabolites. Based on the results of metabolic pathway enrichment and previous research, it is speculated that the synthesis of photosynthetic bacterial carotenoids uses acetyl-CoA as a precursor substance, which is catalyzed by HMG-CoA reductase to synthesize IPP and participate in the synthesis of carotenoids (Helen et al., 2008). The data acquired in this study will facilitate future functional studies of genes/pathways associated with metabolic reactions during the photosynthetic bacterial carotenoid synthesis pathway. The metabolism of microorganisms is a complex process, and deeper and more detailed research is needed in the future on the bacterial carotenoid synthesis pathway.