ABSTRACT
Pseudomonas aeruginosa (P. aeruginosa) has been found to be a common hospital acquired pathogen, responsible for several severe infections. The objective of this study was to describe the antibiotic resistance profile of P. aeruginosa strains from human sample. This descriptive study was carried out on 168 isolated strains of P. aeruginosa collected from January 2014 to December 2015 at the Pasteur Institute of Côte d'Ivoire. The strains identification was done according to the methods of conventional bacteriology. The antibiotic sensitivity tests were performed using the disc diffusion method in agar medium according to CA-SFM (Antibiogram committee of French society of microbiology) criteria. The serotyping of the strains was carried out by using the agglutination method slide test, with the aid of 4 specific polyvalent antisera. The most prevalent P. aeruginosa serotypes were O4 (24.4); O11 (14.6); and O6 (9.5%). The rate of antibiotic resistance to ticarcillin was 32.9%, ciprofloxacin 18.4%, ceftazidime 14.9%, Imipenem 11.3%, and amikacin 11.3%. Resistance to Imipenem was above 10% in an intensive care unit and in the pneumonology unit (PPH). Strains of O6 serotypes were the most multidrug-resistant followed by O11 with respective rates of 31.2 and 28% MDR (Multidrug Resistance). P. aeruginsa are microorganism capable of developing mechanisms of complex resistance which makes it difficult to manage. The attention of hygiene rules and the rational use of antibiotics are very important in other to prevent the spread of MDR P. aeruginosa.
Key words: Pseudomonas aeruginosa, serotype, multidrug-resistance, infection.
Pseudomonas aeruginosa has become a major hospital acquired pathogen, responsible for several severe infections (Bertrand et al., 2013). Due to its ability to use different organic compounds as energy substrates, this strictly aerobic gram-negative bacillus lives in very humid environments (Fuentefria et al., 2011).
Hospital environment is suitable for this micro-organism because it contains numerous ecological niches (Bertrou et al., 2000). The intestinal carriage of the pyocyanic is rare in healthy subjects, but frequent among hospitalized patients, particularly in intensive care unit (Bertrand et al., 2013). Due to its natural resistance to many antibiotics and its ability to build up new resistance, the hospital environment favors the growth of P. aeruginosa despite multiple antibiotic selection pressure.
The emergence of multidrug-resistant strains of P. aeruginosa in hospital is often associated with some serotypes (Thrane et al., 2015). In order to find an epidemiological link between the strains of P. aeruginosa isolated in hospital, phenotypic markers such as antibiotype and serotype are used to differentiate strains before genetic characterization (Blot et al., 2013; Wolska et al., 2012). There is paucity of data on P.aeruginosa infections in Côted’ Ivoire. The aim of this study was to evaluate the antibiotic resistance profile of P. aeruginosa and to compare these profiles with serotypes of isolated strains.
This descriptive study was carried out on strains P. aeruginosa, isolated and collected from January 2014 to December 2015 at the Clinical Bacteriology Unit (CBU) of the Department of Bacteriology-Virology of Pasteur Institute of Côte d'Ivoire. The variables taken into account were: the origin of the strains, type of services, nature of the samples, antibiotic profile and the type of the serotype. Duplicates were not considered.
Strains of P. aeruginosa
A total of 168 strains of P. aeruginosa were isolated from various biological fluids: pus (87), sputum (320), urine (16), blood (13), material (13), Cerebrospinal fluid (6) and stools (1). Strains isolated from other media were examined for growth and pigmentation on Pseudomonas isolation agar. The strains were identified based on the standard bacteriological characteristics: appearance of colonies on King A and King B medium, presence for pyocyanine and pyoverdine, growth at 42°C, morphological appearance (gram negative bacillus, polar mobile), the positive oxidase reaction and other biochemical characteristics using the API 20 NE gallery (Bio-Mérieux®, France) according to Breed et al. (2000) and Garrity et al. (2005). The strains were stored in deep agar at room temperature.
Study of antibiotic sensitivity
Antibiotic susceptibility tests were performed using Kirby-Bauer disc diffusion method in agar medium (Kirby-Bauer, 1996). The reading and interpretation were done according to the recommendations of the Antibiogram Committee of the French Society of Microbiology (CA-SFM 2015).
The following antibiotics were tested: ticarcillin (75 μg), ticarcillin + clavulanic acid (75/10 μg), ceftazidime (30 μg), imipenem (10 μg), amikacin (30 μg) and ciprofloxacin (5 μg). The internal quality control was carried out using the reference strain P. aeruginosa ATCC 27853.
Serotyping
The serotyping of the strains was performed using the slide agglutination technique with the aid of 4 polyvalent specific antisera PMA (O1 + O3 + O4 + O6), PMC (O9 + O10 + O13 + O14), PME (O5 + O15 + O16), PMF (O7 + O8 + O11 + O12) (Biorad ®) followed by specific monovalent antisera (Biorad®).
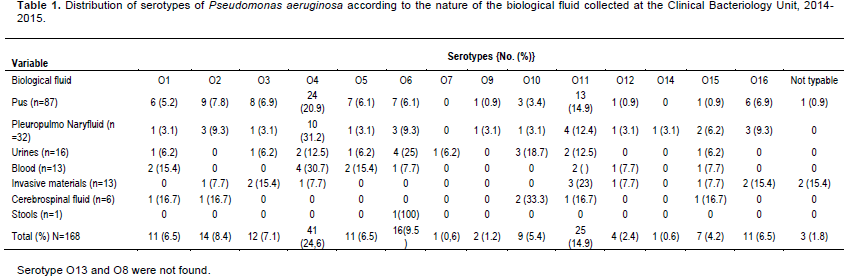
P. aeruginosa were isolated from different biological fluid at variable frequencies, pus (51.8%), followed by pulmonary secretions (19%), urine (9.5%), blood (7.7%), Invasive material (7.7%) and cerebrospinal fluid (3.6%).
Overall, the most prevalent serotypes were O4 (24.4%), O11 (14.6%), and O6 (9.5%). O4 serotype was most frequent in pus with 28%, pulmonary secretions (31%) and urine (30.7%). In urine and hospital materials, O6 and O11 serotypes were the most commonly found (Table 1).
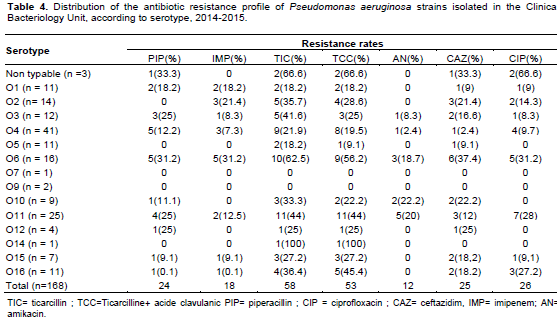
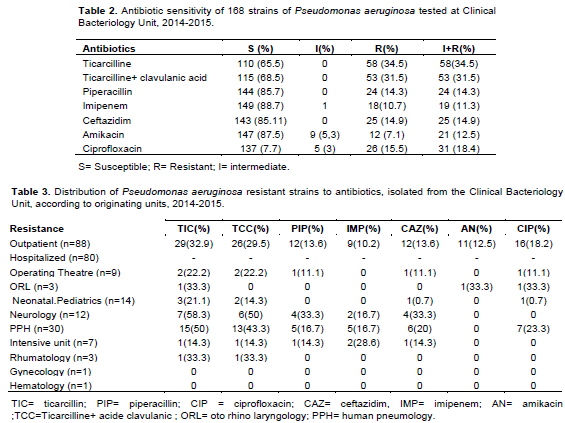
In Table 2, the rate of sensitivity of 168 strains of P. aeruginosa to different antibiotics tested is summarized. The proportion of resistant strains was high for ticarcillin (34.5%) and ticarcillin-clavulanic acid (31.5%). The rate of strains resistant to cetazidime was 14.9 and 18.4% for ciprofloxacin. The resistance rate to Imipenem was 11.3%. Distribution of resistant strains according to their isolation origin showed a higher proportion from hospital strains compared to isolates from non-hospitalized patients. Among hospital services, the highest rates of resistance were observed in pneumonology unit and neurology for ticarcillin (50 and 58.3%, respectively) and ticarcillin-clavulanic acid (43.3 and 50%, respectively). In Intensive care unit, 28.6% is from resistant strain to imipenem. About 23% of strains isolated from pneumonology were resistant to ciprofloxacin (Table 3).
Serotypes O6, O11 and O2 were most associated with resistance to antibiotics. Serotype O6 strain has resistance rate of ticarcillin 62.5%, imipenem 31.2% and ciprofloxacin 37.4% to ceftazidime. For serotype O11, frequent association was observed with ticarcillin resitance (44%), ciprofloxacin resistance (28%) and piperacillin resitance (20%). For serotype O2, the predominant resistance was ticarcillin (35.7%), ceftazidime (21.4%) and imipenem (21.4%) (Table 4).
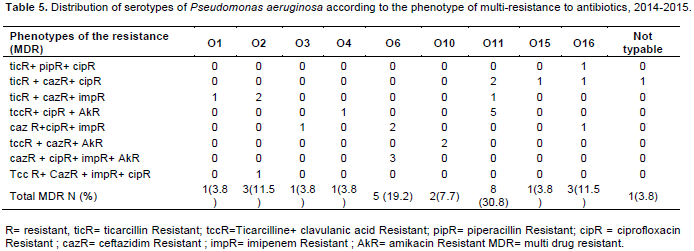
Multiresitance is associated with serotypes O11 (30.8%), O6 (19.2%), O16 (11.5%) and O2 (11.5%). The most common phenotypes were associated to ticarcallin, ceftazidime and ciprofloxacin resistance (Table 5).
P. aeruginosa is generally isolated from suppurative collections as reported by Ben Abdallah et al. (2008) with the rate of 52.9% closer to 51.8%. The presence in other biological fluid is reported with variable proportions 19% in pleuropulmonary secretions and, 7.5% in urine and blood, respectively.
Authors reported 37.2% pus, 19.9% lungs and 30.7% urine (Mindolli et al., 2015) and the rate of 38.5% urine, 8.3% blood and 3.1% catheter (Kiana et al., 2016). The contamination of wounds by this bacterium is usual, however the use of invasive materials in particular the urinary catheters and catheters of intubation could explain their presence in a pleuropulmonary secretions and urines. The rates of P. aeruginosa isolated from hospitalized patients (47.6%) vs. outpatients (52.4%) could be a bias in recruitment, where the previous idea of hospitalization and invasive gestures are not well informed. In Morocco in 2002, the rate was 64.5% among inpatients (Louzi et al., 2003) while in Nigeria in 2012, it was 70% from hospitalized patients (Iregbu et al., 2015). The hospital service most frequently associated is the intensive care unit as reported by several authors (Tacconeli et al., 2002 ; Goncalves et al.,2017). Other services such as pneumology and neurology are also at-risk (Iregbu et al., 2015).
The serotypes of P. aeruginosa constitute epidemiological markers with the rate of 24.4, 14.9 and 9.5% for serotypes O4, O11 and O6, respectively. In Tunisia, a study carried out in 2012 reported the same serotypes with variable rates predominance of serotypes O11, followed by O6 and O4 (Zoghlami et al., 2012). Serotype O4 was predominant in prolong hospitalization units (19.8%) (Adjerald et al., 1999).
Regarding the resistance to antibiotics, the higher rates were recorded for Ticarcillin (34.5%), ciprofloxacin (15.5%) and ceftazidime (14.9%). In Algeria, similar rates were observed for ticarcillin (32.7%), ciprofloxacin (19.4%) and ceftazidime (14.7%) (Sefraoui, 2015) while In Nigeria in 2015, higher rates were reported for ceftazidime (46%) and ciprofloxacin (34%) (Iregbu et al., 2015). In Côte d'Ivoire, a study carried out in 2013 on hospital effluents showed very high rates of resistance to ceftazidime and ticarcillin, resistant strains of 100% (Guessennd et al., 2013).
Amikacin and imipenem were the most active antimicrobial agent on P. aeruginosa isolates with resistance rates of 7.1 and 10.7%, respectively. Nigeria resistance rates were 13% for imipenem and 23% for amikacin (Iregbu et al., 2015).
In Côte d'Ivoire in 2008, P. aeruginosa isolated from infections which originate from surgical operation site shown a sensitivity of 98.5% to an imipenem, that is 1.5% resistance with lower rate (Faye-Kette et al., 2008) . A study carried out in 2013 on hospital effluents showed very high rates of resistance to imipemen with 80% of resistant strains (Guessennd et al., 2013).In Teheran, higher rate were found for imipenem (41.3%) and amikacin (28%) (Peymani et al., 2017).
The highest resistances were observed in strains isolated from the department of pulmonology with 50% resistance for ticarcillin, 23.3% for ciprofloxacin and 20% for ceftazidime. This observation is confirmed by Hamze et al. (2013) in Lebanon.
The serotypes O11 (30.8%) and O6 (19.2%) presented more multiresistant strains. Association of serotypes and multiresistance to antibiotics was reported by several studies, as in the case with serotype O11 (Sardelic et al., 2012). Approximately, 32% of the multi-resistant strains of this study belonged to serotype O11. Similarly, in Greece, serogroups O12 and O11 had multiresistance rates of 91 and 79%, respectively (Panayotis et al., 1998), whereas in Japan in 2007, out of 214 strains of P. aeruginosa of the serotype O11, 212 (99%) were MDR (Jun-Ichiro et al., 2007).
This study showed the importance of serotyping of P. aeruginosa isolated from clinical sources. The circulation of serotypes O4, O11 and O6 of P. aeruginosa was frequent. They were predominant in suppurative and pleuro-pulmonary secretions. In addition, serotypes O11 and O6 were the most multiresistant. The most common phenotype of MDR is Ticarcillin-Ceftazidime-Ciprofloxacin. Knowledge of serotypes can guide the choice of antibiotic therapy in 24 h before sensitivity test results. In addition, this study should alert health professionals to an increasing rate of P. aeruginosa resistant to useful carbapenems and Fluoroquinolones.
Regular monitoring of the antimicrobial resistance profile is essential to guide prescribing antibiotics and controlling the emergence of MDR P. aeruginosa strains.
The authors have not declared any conflict of interests.
REFERENCES
|
Adjerald L, Cattoen C, Levent T, Grandbastien B, Descamps D, Bouillet L, Coignard B, Beaucaire G (1999). Réseau de microbiologistes de l'ARECLIN. Observatoire régional Pseudomonas aeruginosa du Nord-Pas-de-Calais: Données épidémiologiques et microbiologiques. Med Mal Infect; 29(3):160-166.
Crossref
|
|
|
|
Bertrou A, Chapuis C, Hajjar J (2000). Relation entre contamination et environnement hospitalier. In vigilance environnementale: Contrôles microbiologique de l'environnement hospitalier. Hygiène 8 (3):143-146.
|
|
|
|
Blot S, Koulenti D, Dimopoulos G, Martin C, Komnos A, Krueger WA, Spina G, Armaganidis A, Rello J (2013). Prevalence, risk factors, and mortality for ventilator-associated pneumonia in middle-aged, old, and very old critically Ill patients. Crit. Care Med. (2014) Mar; 42(3):601-9
|
|
|
|
Breed Rs, Murray EGD, Smith Nr (2000). Bergey's Manual of Determinative Bacteriology, seventh edition, the Williams & Wilkins Co, Baltimore, 67p.
|
|
|
|
Faye-Ketté H, Kouassi M. Y, Akoua-Koffi G, Bakayoko S, Boni-Cissé C, Diallo-Touré K, Dosso M, Lambin Y (2008). Epidémiologie microbienne des infections de sites opératoires dans un service de traumatologie à Abidjan et sensibilité des germes aux antibiotiques. Revue Bio-Africa - N°6, pp. 25-31.
|
|
|
|
Fuentefria DB, Ferreira AE, Corcao G (2011). Antibioticresistant Pseudomonas aeruginosa from hospital wastewater and superficial water: Are they genetically related. J Environ Manage. 92:250.
Crossref
|
|
|
|
Garrity Gm, Bell Ja, Lilburn T. (2005). Bergey's Manual of Systematic Bacteriology, second edition, vol. 2 (The Proteobacteria), part B (The Grammaproteobacteria), Springer-Verlag, New York, 323p
|
|
|
|
Goncalves I R, Dantas RCC, Ferreira ML, Deivid, Batistão DWF, Gontijo-Filho PP, Ribas RM (2017). Carbapenem-resistant Pseudomonas aeruginosa: association with virulence genes and biofilm formation. Braz j. microbiol. 4(8):211-217.
Crossref
|
|
|
|
Guessennd NK, Ouattara MB, Ouattara ND, Nevry RK, Gbonon V, Tiekoura KB, Dosso M (2013). Étude des bactéries multi résistantes des effluents hospitaliers d'un centre hospitalier et universitaire (CHU) de la ville d'Abidjan (Côte d'Ivoire). J. Appl. Biosci. 69:5456- 5464.
Crossref
|
|
|
|
Hamze M, Mallat H, Dabboussi F, Achkar M (2013). Antibiotic susceptibility and serotyping of clinical Pseudomonas aeruginosa isolates in northern lebanon. Int. Arab. J. Antimicrob. Agents 4(2):720.
|
|
|
|
Iregbu KC, Eze SO (2015). Pseudomonas aeruginosa infections in a tertiary hospital in Nigeria. Afr. J. Cln. Exper. Microbiol. 16(1):33-36.
Crossref
|
|
|
|
Jun-Ichiro S, Tsukasa A, Miyoshi-Akiyama T, Atsushi K, Yukie M, Minako A, Tomoko F, Hideko K, Satoru S, Hajime W, Tadashi K,Hiroshi M, Keiji K, Hiroyuki K, Yoshihiro K, Mitsuo K, Hiroshi Y, Tadatoshi K and Teruo K (2007). Outbreaks of Multidrug-Resistant Pseudomonas aeruginosa in Community Hospitals in Japan. J. Clin. Microbiol. 45(3):979-989.
Crossref
|
|
|
|
Kiana S, Behrouz A, Fardad (2016). Antibiotic resistance pattern and evaluation of metallo-beta lactamase genes (VIM and IMP) in Pseudomonas aeruginosa strains producing MBL enzyme, isolated from patients with secondary immunodeficiency. Adv. Biomed. Res. 5:124.
Crossref
|
|
|
|
Kirby-Bauer A. (1996) Antimicrobial sensitivity testing by agar diffusion method. J. Clin. Pathol. 44:493.
|
|
|
|
Louzi L, Boughalem M, Charra B, Jana M. (2003) Pseudomonas aeruginosa: profils de resistance aux antibiotiques à propos de 62 souches. J. magh. A. Réa. 2001-2002 vol x-P. 191.
|
|
|
|
Mindolli PB, Salami MP (2015); Antimicrobial susceptibility pattern of Pseudomonas aeruginosa from clinical isolates at a tertiary care center of in jivaypur karna. J. Chem. Phama. Res. 7(8):186-190.
|
|
|
|
Tassios PT, Gennimata V, Maniatis AN, Fock C, Legakis NJ (1998). Greek Pseudomonas aeruginosa Study Group. Emergence of multidrug resistance in ubiquitous and dominant Pseudomonas aeruginosa serogroup O: 11. J. Clin. Microbiol. 36(4):897-901.
|
|
|
|
Peymani A, Naserpour-Farivar T, Zare E, Azarhoosh Kh. (2017). Distribution of blaTEM, blaSHV, and blaCTX-M genes among ESBL-producing P. aeruginosa isolated from Qazvin and Tehran hospitals, Iran. J. Prev. Med. Hyg. 58:E155-E160.
|
|
|
|
Mindolli PB, Salami MP (2015). Antimicrobial susceptibility pattern of Pseudomonas aeruginosa from clinical isolates at a tertiary care centre of in jivaypur karna. J. Chem. Phama. Res. 7(8):186-190.
|
|
|
|
Rana Barakat (2012). Etude des propriétés biologiques et antimicrobiennes de la pyocyanine, pigment redox-actif produit par Pseudomonas aeruginosa. Sciences agricoles. Université de La Rochelle, Disponible :
View
|
|
|
|
Sanda S, Branka B, Colinon-Dupuich C, Orhanovic S, Zrinka B, Plecko V, Cournoyer B and Gian M.R (2012). Infrequent Finding of Metallo-β-Lactamase VIM-2 in Carbapenem-Resistant Pseudomonas aeruginosa Strains from Croatia. Antimicrob Agents Chemother; 56(5):2746-2749.
Crossref
|
|
|
|
Sefraoui Imane Epse Khelil (2015). Etude de la résistance aux antibiotiques de Pseudomonas aeruginosa au niveau de différents hôpitaux de l'Ouest algérien. Thèse de doctorat en Biologie présentée et soutenue le 29 janvier 2015.
|
|
|
|
Tacconelli E, Tumbarello M, Bertagnolio S, Citton R, Spanu T, Fadda G (2002). Multidrug-resistant Pseudomonas aeruginosa bloodstream infections: analysis of trends in prevalence and epidemiology. Emerg Infect Dis. 8(2):220-221.
Crossref
|
|
|
|
Valerie A, Navon-Venezia S, Seigman-Igra Y, Shaltiel C, and Yehuda C (2006). Multidrug-Resistant Pseudomonas aeruginosa: Risk Factors and Clinical Impact. Antimicrob Agents Chemother. 50(1):43-48.
Crossref
|
|
|
|
Xavier B, Céline S, Pascal C, Talon D (2011). Épidémiologie des infections à Pseudomonas aeruginosa. Revue Francophone des Laboratoires. 435:35-40.
|
|
|
|
Zoghlami A, Kanzari L, Boukadida J, Messadi AA, Ghanem A (2012). Epidemiological profile and antibiotic resistance of Pseudomonas aeruginosa isolates in burn and Traumatology center in Tunisia over a three-year period. Tunis Med. 90 (11):803-806.
|
|
|
|
Thrane SW, Taylor VL, Freschi L, Kukavica-Ibrulj I, Boyle B, Laroche J, Pirnay JP, Lévesque RC, Lam JS, Jelsbak L (2015). The widespread multidrug-resistant serotype O12 Pseudomonas aeruginosa clone emerged through concomitant horizontal transfer of serotype antigen and antibiotic resistance gene clusters. MBio. 6(5):e01396-15.
Crossref
|
|
|
|
Wolska K, Kot B, Jakubczak A (2012). Phenotypic and genotypic diversity of Pseudomonas aeruginosa strains isolated from hospitals in Siedlce (Poland). Brazilian J. Microbiol. 43(1):274-282.
Crossref
|