ABSTRACT
Many soils of the inter-tropical regions are P-deficient because of their high fixing power and low P content. Rock phosphate resources used to produce the phosphate fertilizers are exhausted and chemical fertilizer are causing environmental degradation. This issue raised the question of sustainability of fertilization and subsequently has enhanced the interest in the use of microorganisms as biofertilizers. The aim of this study is to isolate and characterize potential P solubilizing bacteria (PSB) from two P deficient agricultural regions in Senegal. Twelve potential PSB were selected and further screened for other plant growth promoting traits (Indole-3-acetic acid (auxin) and siderophore production) and characterized by 16S rDNA sequencing. All the isolates produced auxin and seven of them produced siderophore. DNA sequencing showed that five isolates were affiliated to the genus Bacillus, four to the genus Staphylococcus, two to the genus Microbacterium and one isolate showed high similarities with members of the genus Burkholderia. The selected bacteria will further be tested on some plants to assess their biofertilization potential.
Key words: 16S rDNA, indole-3-acetic-acid (IAA), phosphate solubilizing bacteria (PSB), siderophore.
Early studies proved the existence of a group of soil free living bacteria stimulating plant growth, which was called plant growth promoting rhizobacteria (PGPR) (Kloepper and Schroth, 1978). Since, then, mechanisms for stimulating plant growth were described. PGPR may produce various compounds, including growth regulators (phytohormones), siderophores, and organic acids. Some are able to ï¬x atmospheric nitrogen, solubilize phosphorus and produce antibiotics to suppress harmful rhizobacteria (Sureshbabu et al., 2016). These mechanisms either directly affect the metabolism of plants or improve the adaptive capacity of plants to acquire other nutrients from the soil (Santoro et al., 2015). PGPR that are able to mineralize soil phosphorus are called phosphate solubilizing bacteria (PSB). Phosphate solubilization is considered to be most important attribute of plant growth promoting rhizobacteria (Kloepper et al., 1989).
Phosphorus (P) plays an important role in plant physiology and is frequently the prime limiting factor for plant growth in terrestrial ecosystems (Bunemann et al., 2011). Phosphate is the second most important element for mineral nutrition of plants and by far the least mobile and available to plants in most soil conditions (Hinsinger, 2001). Phosphate is present in the soil at levels of 400 to 1,200 mg kg-1; however, soluble P concentrations in soil are generally very low, at levels of 1 mg kg-1 or less (Goldstein, 1994). The poor availability of soil inorganic phosphate is due to the large reactivity and retention of these phosphate ions with other metals (Fe, Al, Ca) (Rodriguez and Fraga, 1999; Hinsinger, 2001).
Application of P-containing fertilizers is common for stimulating crop yields. However, repeated applications of phosphate fertilizers affect environment, microbial diversity and can lead to loss of soil fertility and consequently lower crop yields (Gyaneshwar et al., 2002). Thus, it is a great challenge to search for strategies that may alleviate detrimental effects of current intensive farming practices that use chemical fertilizers. An attractive alternative to the phosphatic fertilizers is the use of PSB as biofertilizer that have been shown to enhance plant growth and improve P availability in the soils (Pereira and Castro, 2014). Nevertheless, plant growth enhancement may also be related to other PGP traits that may act in synergy with P solubilization like indol-3-acetic acid and siderophore production (Pereira and Castro, 2014). Moreover, plant growth enhancement seems to be related not only to P solubilization but also to other PGP traits, like indol-3-acetic acid and siderophore (Pereira and Castro, 2014). Inoculation success also is related to the persistence of the introduced strain, that is, its ability to establish high population levels and to live as a continuing member of the soil microflora even in the absence of plant (Lupwayi et al., 2006). Introduced bacteria are not always competitive with native soil microbial communities (Herrmann and Lesueur, 2013), as they have to compete for niches and nutrients in new environmental conditions.
With the aim to develop a biofertilizer adapted to soils in major cultivation areas of Senegal, PGPR were isolated from P-deï¬cient soils in two agricultural regions (Kaffrine and Kolda). PSB were first isolated from rhizosphere soil and from non-rhizospheric soils, screened for other plant growth promoting traits (indole-3-acetic acid (IAA) and siderophore production) and characterized at the molecular level by 16S rDNA sequencing.
Soil sampling
Rhizospheric soils and non rhizhospheric soil (bare soil) were collected in Senegal from two sites (Kolda: 12°50’N - 14°50’W and Kaffrine: 13° 57’N- 15° 35’W) (Figure 1). Bare soils were sampled from the top 20 cm soil free of litter. Rhizospheric soils were sampled from roots of Guiera senegalensis, Piliostigma reticulatum and Dichrostachys glomerata. Roots with adhering soil were put in a plastic bag, shaken with hands for 5 min to collect rhizospheric soil and removed. Seventy-six (76) samples were carefully collected in bags and stored at 4°C temperature for the isolation of bacterial strains (PSB). Table 1 indicates the P level in collected soils, showing that even total P content is low.
Isolation and characterization of PSB
For isolation of PSB, 10 g soil samples were suspended in 90 ml of NaCl buffer. A serial dilution assay was carried out in 0.9% NaCl buffer (NaCl: 4.38 g/500 ml, KH2PO4: 0.135 g/500 ml; Na2HPO4+ 2H2O or Na2HPO4: 0.284 g/500 ml) solution. An aliquot of 0.1 ml of each dilution (10-2, 10-3 and 10-4) was spread onto Petri plates containing Pikovskaya (PKV) agar (Pikovskaya, 1948). The composition of PKV medium was (g.l-1): glucose: 10.0; Ca3(PO4)2: 5; (NH4)2SO4: 0.50; KCl: 0.20; Mg2SO4ï½¥7H2O: 0.010; Mn2SO4ï½¥H2O: 0.0001; Fe2SO4ï½¥7H2O: 0.0001, yeast extract: 0.50; pH was adjusted to 7.0. The plates were incubated at room temperature (28°C) for 7 days. Colonies showing a clear zone around the colony was considered as P-solubilizer. The P-solubilizers were purified by repeated streaking and stocked for further use.
Biochemical characterization of PSB isolates
Phosphate solubilization
An aliquot of 0.1 ml of each PSB culture preserved was placed on Pikovskaya’s agar (PA) (Petri dish) (Pikovskaya, 1948). The plates were incubated at room temperature (28°C) for 7 days. The solubilization zone was determined by subtracting the diameter of bacterial colony from the diameter of total zone. Solubilization index (SI) was calculated using the formula:
Solubilization of tri-calcium phosphate was quantified in Pikovskaya’s broth. Each flask containing 75 ml medium (PVK) was inoculated with 500 µl of bacterial culture (three replicates were performed for each isolate) and incubated at 28 ± 0.1°C at 140 revolutions per minute (rev.min-1) for 4 days in incubator. Simultaneously, a non-inoculated control (free PVK medium) was also kept under similar conditions. Cultures were harvested by centrifugation at 13,000 g for 10 min. The soluble P expressed as mg.l-1 in bacterial isolates was quantified by the colorimetric method of Olsen and Sommers (1982).
Screening of indole-3-acetic acid (IAA)-producing bacteria
For detection of IAA production by PSB strains, Luria-Bertani solid medium (LBT) enriched with L-tryptophan (1 g l-1) was prepared and flowed into the dishes (Bric et al., 1991). A nitrocellulose membrane was placed directly on the LBT medium and inoculated with the isolates using a loop. The Petri dishes were then incubated at 28°C, for 2 to 4 days (the time required for the colonies to reach a diameter of 2 mm). A 9 cm Whatman qualitative filter paper (No. 2) was impregnated with 2.5 ml of Salkowski's solution (30.8 ml of water, 19.3 ml of 96% pure H2SO4 sulfuric acid and 0.6 g of trichloride of iron) (Gordon and Weber, 1951). The nitrocellulose membrane, showing growths of colonies were dropped on a filter paper impregnated with Salkowski's reagent. Bacteria that synthesized IAA were identified by the formation of a characteristic red halo that surrounds the colony.
The production of IAA was quantified following the method of Salkowski (Gravel et al., 2007). The isolates were first cultured in Tryptic Soy Broth (TSB) for 24 h at 28°C, then 30 µl of the pure culture was inoculated into test tubes containing 3 ml of Luria Bertani (LB) medium supplemented with 1 g.l-1 L- tryptophan. The test tubes containing the bacterial isolates were incubated for 5 days with shaking (200 rev.min-1) at 28°C. The determination of the IAA concentration was carried out by the addition of 100 µl of the Salkowski solution [30.8 ml water, 19.3 ml of pure sulfuric acid H2SO4 96% and 0.6 g of Iron (III) chloride] (Gordon and Weber, 1951)to 100 μl of the culture supernatant which had been previously centrifuged for 20 min at 13,000 g. After 20 min incubation at room temperature, the optical density at 535 nm was recorded. IAA production is indicated by the presence of a pinkish color. The following dilutions: 10, 25; 100, 200 and 400 ng.ml-1 of IAA (Sigma I-2886) was used to establish a standard curve (r2 = 0.9997). This range of dilutions was prepared from a 10-3 M auxin stock solution by diluting 17.5 mg of auxin in 1 ml of absolute ethanol and then adjusting the volume to 100 ml with sterile demineralized water (Gupta et al., 2014). The amount of IAA produced was expressed as ng.ml-1 by comparison with the standard curve.
Siderophore assay
The production of siderophore in liquid and solid medium was tested in King B medium and the Chrome azurol-S (CAS) following the methodology described by Schwyn and Neilands (1987)and modified by Milagres et al. (1999). The King B medium was prepared and mixed with CAS in the following proportions: 100 ml of CAS + 900 ml of King B. After solidification, the plates were inoculated with the pure culture of bacteria (30 µl) and incubated at 28°C for 4 days. The presence of an orange-yellow halo around the strain is described as positive for the production of siderophore. This color change is due to the transfer of ferric ions from the CAS to the siderophore.
The siderophore production of isolates was also tested by the method of Ribeiro and Cardoso (2012). This involves inoculating tubes containing 3 ml of liquid KB medium with isolates and incubating for 7 days at 28°C under constant agitation. After this period, 1 ml of the culture was added to 1.5 ml microtubes and centrifuged for 5 min at 14,000 g. Then 100 µl of the supernatant from each culture was added to a microplate well containing 100 µl from the reagent chrome azurol S (CAS) and incubated for 30 min. The orange or yellow coloring of the medium indicates the production of siderophores by bacteria. The uninoculated KB medium was used as negative control. Absorbance was read at 630 nm. The following equation was used to calculate the percent activity of siderophore produced.
Molecular identification of PSB
DNA extraction
The identification of PSB was done by 16S rDNA gene sequencing. The genomic DNA of PSB was extracted by the E.Z.N.A. Bacterial DNA kit of OMEGA bio-tek (400 Pinnacle Way Suite 450 Norcross, GA, 30071 USA) was according to instructions of the manufacturer.
PCR amplification of bacterial 16S rDNA
The variable region (V3) of the 16S rDNA gene was targeted for the identification of PSB bacterial strains. The gene encoding the 16S subunit ribosomal DNA (rDNA) is mostly used because of its structure highly conserved in all bacteria which is very useful for the identification of universal primers (Hillis and Dixon, 1991). Universal bacterial primers fD1 (5'-AGAGTTTGATCCTGGCTCAG-3') and rD1 (5'- AAGGAGGTGATCCAGCCGCA-3') (Weisburg et al., 1991)were used for amplification of the V3 region. The PCR reaction mixture was composed of 0.25 μl dNTP (10 mM each dNTP), 2.5 μl MgCl2 buffer (25 mM), 0.22 μl GoTaq (X5), 5 μl GoTaq buffer (5 U/µl), 1 μl of the DNA sample, 1.25 μl (of each) Primers (dD1 and rD1 at 10 μM) and sterile H2O to reach 25 μl (final volume per sample). The following cycling conditions were used: a first denaturation phase (5 min at 94°C) was followed by 35 cycles of 94°C for 30 s (denaturation), 55°C for 1 min (Hybridization) and 72°C for 1 min (Elongation), and finally a final elongation step at 72°C for 5 min. The PCR amplification was carried out using a Gene Amp PCR System 9700 thermocycler (Applied Biosystem).
Sequencing and phylogeny analyses
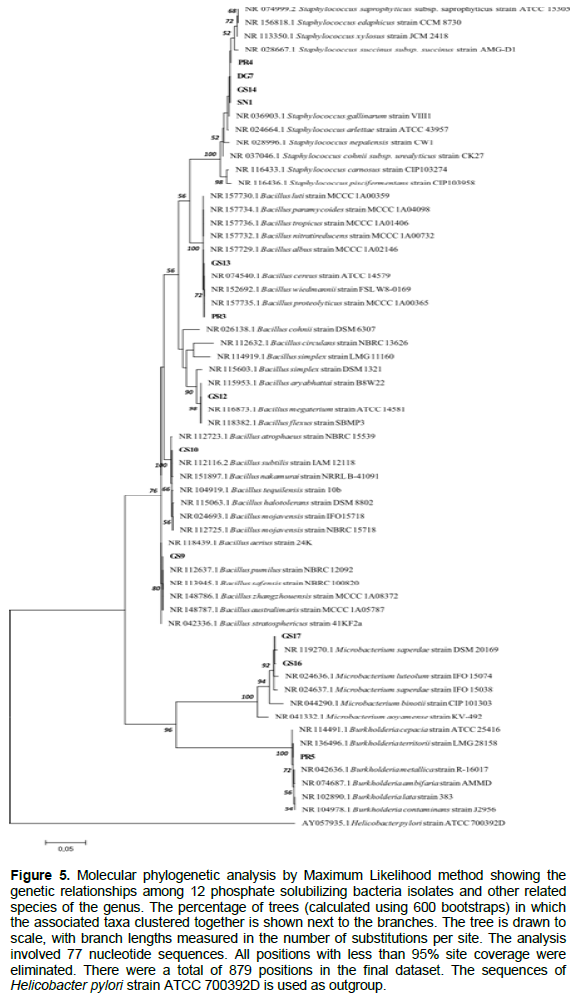
The PCR products were sequenced by GENEWIZ (USA). The sequences were compared with National Center of Biotechnology Information-USA (NCBI) database using BLAST method. The evolutionary history was inferred by the Maximum Likelihood method based on the Kimura 2-parameter model with 600 bootstraps. Phylogenetic analysis was conducted using MEGA 7.0.
Statistical analysis
Determination of significant differences between strains for quantitative PGP production was performed by using one-way analysis of variance (ANOVA) and non-parametric test. Fisher’s and Kruskal Wallis’s paired multiple comparison post-hoc tests with the software XLSTAT (XLSTAT 2016 Addinsoft, France) was carried out if the difference between the treatments was significant. Least significant differences (LSD) were calculated at the 5% level.
Selection of PSB and biochemical characterization of PS activity
PSB isolation and solubilization index (SI)
After 7 days of incubation at room temperature (28°C), bacterial isolates producing transparent halos on PVK solid medium were considered as PSB. A total of 12 strains solubilizing phosphate were obtained (Table 2). One strain was obtained from non-rhizospheric soil (SN1) and the other 11 from rhizospheric soil of the followed species: P. reticulatum (PR3, PR4 and PR5), G. senegalensis (GS9, GS10, GS12, GS13, GS14, GS16 and GS17) and D. glomerata (DG7).
The SI determined ranged from 11.61 to 21.40 mm between the isolates (Table 2). Eleven (11) isolates exhibit a SI higher than 15 mm. The lowest SI was recorded in isolate GS10 with 11.61 mm. The highest SI was found with GS17 (21.40 mm), DG7 (20.40 mm) and GS14 (20.35 mm).
P-solubilization in liquid culture
When cultivated in liquid PVK medium, strains showed high variation for their ability to solubilize tricalcium phosphate, from 53.54 to 423.41 mg.l-1 (Figure 2). The isolate GS17 originating from rhizosphere soil of G. senegalensis in Kaffrine solubilized significantly higher phosphate than all other bacterial strains. The lowest solubilization capacity was obtained from GS9 (53.54 mg.l-1). The solubilization capacity was not correlated to the origin of the strain.
IAA production
All the bacterial isolates induced a red halo surrounding the colony, showing IAA activity (Table 2). Quantitative estimation of IAA production was made by comparison with standard curve. Production of IAA by the isolates ranged from 47.94 to 575.87 µg.ml-1 (Figure 3). The highest concentration (248.02 µg.ml-1) was produced by the isolate GS12 in liquid LB broth (Figure 3). The lowest IAA production was shown by PR3, GS16, DG7 and GS17.
Siderophore production
Incubation in King B and Chrome-azurol-S (CAS) solid medium showed seven isolates with yellow coloring both in solid and liquid medium, demonstrating their capacities to produce siderophores (Figure 4 and Table 2). Quantitative estimation of siderophore using Chrome-azurol-S (CAS) liquid assay revealed that SN1 is the highest siderophore rate (36.19%) (Figure 4). Minimum siderophore production (0.17 and 0.23%) was found in isolates GS14 and GS9.
Molecular identification
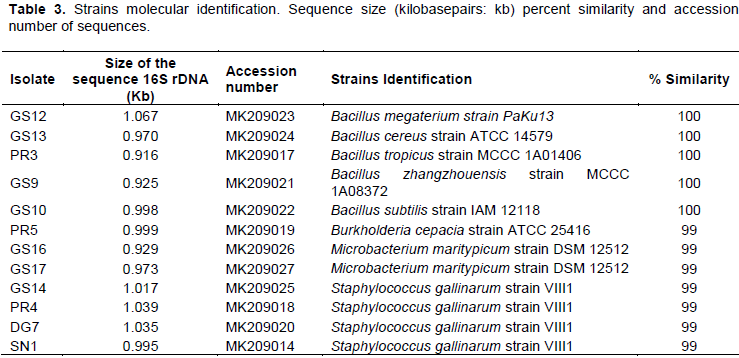
Analysis of 16S rDNA gene sequences using data available in GenBank showed that five strains belong to genus Bacillus, four to genus Staphylococcus, two to genus Microbacterium and one to genus Burkholderia (Table 3 and Figure 5). Among the seven PSB isolated from G. senegalensis rhizosphere in Kaffrine region, four were Bacillus species, two isolates presenting a single nucleotide difference clustered with Microbacterium maritypicum. The remaining isolate (GS14) presented 100% sequence similarities to strains isolated from Kolda either from non rhizospheric soil (SN1) or from the rhizosphere of D. glomerata (DG7) and showed two base differences with one strain isolated from P. reticulatum rhizosphere (PR4); these four strains presented either 1 or 2 base substitutions with Staphylococcus gallinarum strain VIII1. Finally, the two remaining Kolda isolates recovered from the rhizosphere of P. reticulatum were clustering either with several Bacillus spp. including B. cereus with which they shared 100% sequence similarities (PR3) or with a beta-proteobacterium belonging to the genus Burkhloderia (PR5).
Origin and identification of PSB
This study is the first step of a project aiming to develop PGPR based biofertilizers adapted to main agricultural regions of Senegal. Our approach is based on a primary isolation of P solubilizing bacteria from P-deficient soils, in two main agricultural regions of Senegal (Kolda and Kaffrine). Thus, the entry point was the capacity of bacteria for P solubilization using TCP-based medium that were further analyzed for other PGPR traits. Most PSB were isolated from rhizospheric soil of shrubs found locally (G. senegalensis in Kaffrine; P. reticulatum, D. glomerata in Kolda). This result confirms those of de Abreu et al. (2017), which showed that PSBs are ubiquitous in soils. Of the 12 strains isolated in different areas, 11 were isolated in rhizospheric soil and only one was recovered from non-rhizospheric soil. This result suggests that rhizospheric soils are more likely to harbor PSB than non-rhizospheric or bulk soil. Similar results were reported by Baliah et al. (2016)who found abundant populations of PSB in rhizosphere soil compared to non rhizospheric soil. Indeed, the secretion of carbohydrates and amino acids from roots enhances the growth and multiplication of bacterial species and constitutes a biotope suitable for microorganisms growth (Bertin et al., 2003). Phosphate solubilizing bacteria are known to be abundant in the rhizospheric soils of various plants (Ashok et al., 2012), but their presence varied considerably according to plant species (Reyes et al., 2007). According to Marschner et al. (2004), abundance and diversity of microorganisms in the rhizosphere are likely to be related to plant species due to differences in root exudation and rhizodeposition. In this study, more PSB were obtained from G. senegalensis and P. reticulatum rhizospheric soils. These species are known to be very useful in the maintenance of soil fertilization through root exudates and litter inputs (Wezel et al., 2000; Diakhate, 2016).
Strains belonging to the genera Bacillus, Microbacterium, Staphylococcus and Burkholderia were identified in this study. Those strains have already been found to be PSB in other studies (Rodriguez and Fraga, 1999). According to some authors (Bouizgarne, 2013; Kumar et al., 2016), those genera are particularly effective P-solubilizers. In the present study, we found that the majority of isolated strains belong to the genus Bacillus, however, the greatest ability to solubilize phosphate was found in strains belonging to the genus Microbacterium.
Even though many studies reported the isolation of PSB strains using TCP-based medium, it is important to note that, others are questioning the effectiveness of TCP- based medium to assess the capacity of bacteria for P-solubilization (Bashan et al., 2012, 2013). Other media and P sources such as Metal-P (Al-P, Fe-P, Ca-P) are recommended by those authors, depending on the type of soil and the end use of the targeted bacteria: calcium phosphate (including natural phosphates) for alkaline soils, and iron or aluminium phosphates for acidic soils. These screens would probably maximize the chances of selecting the most effective strains able to contribute to the phosphate nutrition of plants but would reduce significantly the number of potential P solubilizing strains, and consequently our chance when exploring other PGRP traits.
PGP characters of strains
In the present study, it was found that the capacity to solubilize inorganic P varied considerably between the isolated strains and was independent of their geographical origin. We also found variations among strains belonging to the same genus or species. Similar results were found on 193 isolates (Solanki et al., 2018)selected from the rhizosphere of chickpea, mustard and wheat showing large variations in P solubilization independently of their origin. Moreover, some authors who worked on Pseudomonas fluorescens strains isolated from various agricultural ï¬elds also indicated that signiï¬cant variations may also exists within a single bacterial species (Browne et al., 2009). To further investigate the potential PGP of isolated strains, IAA and siderophore productions were tested under in vitro conditions.
IAA is a phytohormone produced in large quantities by many PGPR (Vessey, 2003). Some authors report that 80% of rhizobacteria can synthesize IAA (Gupta et al., 2015). In the present study, all selected bacteria had the ability to produce IAA. However, IAA production varied greatly among strains as shown previously by Shahab et al. (2009). According to Walpola and Arunakumara (2016), IAA production by microbial isolates varies greatly among different species and strains and depends on the availability of substrate(s). The production of IAA is also influenced by the culture conditions or the developmental stage (Mirza et al., 2001). Strains belonging to Azospirillum (Dobbelaere et al., 1999), Rhizobium, Microbacterium, Sphingomonas, and Mycobacterium genera (Tsavkelova et al., 2006)are among the most active IAA producers. In this study, high production of IAA was found in a strain belonging to the genus Bacillus (GS12: MK209023), suggesting that other genera may have great potential. Indeed, high IAA production was also found in Bacillus simplex and Paenibacillus polymyxa species (Erturk et al., 2010).
Another important PGP character is the production of siderophore. Siderophore are small organic molecules produced by microorganisms under iron-limiting conditions that enhance iron uptake capacity (Gouda et al., 2018). Iron is essential for the growth of soil microorganisms. The major strategy to acquire iron is the production and utilization of siderophore (Chaiharn et al., 2008). Microbial siderophore enhance iron uptake by plants that are able to recognize the bacterial ferric-siderophore complex (Dimkpa et al., 2008). In this study, seven isolates were positive for siderophore production in King B and CAS solid media. The rhizobacteria that can produce siderophore could compete for iron with soil borne pathogens and may act as biocontrol agents (Chaiharn et al., 2008).
In a context of climate changes, low fertility of soils and an increasing world population, there is a need to develop sustainable agricultural practices. PGPR represent a real option for crop improvement and protection. Here, the first step of a project aiming at developing a biofertilizer constituted of PGPR was achieved. Globally from the 12 isolates studied, seven exhibit high phosphate solubilizing capacity, and produce IAA and siderophore. These represent good candidates for plant growth stimulation. Finally, rhizobacteria that produce siderophores could also compete for iron with soil borne pathogens. These PGPR could also participate in the protection of the plant and thus represent promising biofertilizers adapted to Senegalese soils.
The authors have not declared any conflict of interests.
The authors thank the West Africa Agricultural Productivity Program (WAAPP) and Fonds National de Recherches Agricoles et Agroalimentaires (FNRAA) which supported this work.
REFERENCES
|
De Abreu CS, Figueiredo JEF, Oliveira CA, Dos Santos VL, Gomes EA, Ribeiro VP, Barros B de A, Lana U de P, Marriel IE (2017). Maize endophytic bacteria as mineral phosphate solubilizers. Embrapa Milho E Sorgo-Artigo Em Periód Indexado (ALICE).
Crossref
|
|
|
|
Ashok VG, Sabina SS, Preeti GD (2012). Isolation and identification of phosphate solubilizing fungi from rhizosphere (soil). International Journal of Science Innovations and Discoveries 2:310-315.
|
|
|
|
|
Baliah NT, Pandiarajan G, Kumar BM (2016). Isolation, identification and characterization of phosphate solubilizing bacteria from different crop soils of Srivilliputtur Taluk, Virudhunagar District, Tamil Nadu. Tropical Ecology 57:465-474.
|
|
|
|
|
Bashan Y, Kamnev AA, de-Bashan LE (2012). Tricalcium phosphate is inappropriate as a universal selection factor for isolating and testing phosphate-solubilizing bacteria that enhance plant growth: a proposal for an alternative procedure. Biology and Fertility of Soils 49:465-479.
Crossref
|
|
|
|
|
Bashan Y, Kamnev AA, de-Bashan LE (2013). A proposal for isolating and testing phosphate-solubilizing bacteria that enhance plant growth. Biology and Fertility of Soils pp. 1-2.
Crossref
|
|
|
|
|
Bertin C, Yang X, Weston LA (2003). The role of root exudates and allelochemicals in the rhizosphere. Plant Soil 256:67-83.
Crossref
|
|
|
|
|
Bric JM, Bostock RM, Silverstone SE (1991). Rapid in situ assay for indoleacetic acid production by bacteria immobilized on a nitrocellulose membrane. Applied and Environmental Microbiology 57:535-538.
|
|
|
|
|
Browne P, Rice O, Miller SH, Burke J, Dowling DN, Morrissey JP, O'Gara F (2009). Superior inorganic phosphate solubilization is linked to phylogeny within the Pseudomonas fluorescens complex. Applied Soil Ecology 43:131-138.
Crossref
|
|
|
|
|
Bunemann EK, Oberson A, Frossard E (2011). Phosphorus in Action. Soil Biology 26:37-57.
Crossref
|
|
|
|
|
Chaiharn M, Chunhaleuchanon S, Kozo A, Lumyong S (2008). Screening of rhizobacteria for their plant growth promoting activities. Current Applied Science and Technology 8(1):18-23.
|
|
|
|
|
Diakhate S (2016). Soil microbial functional capacity and diversity in a millet-shrub intercropping system of semi-arid Senegal. Journal of Arid Environments 129:71-79.
Crossref
|
|
|
|
|
Dimkpa C, Svatoš A, Merten D, Büchel G, Kothe E (2008). Hydroxamate siderophores produced by Streptomyces acidiscabies E13 bind nickel and promote growth in cowpea (Vigna unguiculata L.) under nickel stress. Canadian Journal of Microbiology 54:163-172.
Crossref
|
|
|
|
|
Dobbelaere S, Croonenborghs A, Thys A, Broek AV, Vanderleyden J (1999). Phytostimulatory effect of Azospirillum brasilense wild type and mutant strains altered in IAA production on wheat. Plant Soil 212:153-162.
Crossref
|
|
|
|
|
Erturk Y, Ercisli S, Haznedar A, Cakmakci R (2010). Effects of plant growth promoting rhizobacteria (PGPR) on rooting and root growth of kiwifruit (Actinidia deliciosa) stem cuttings. Biological Research 43:91-98.
Crossref
|
|
|
|
|
Goldstein AH (1994). Involvement of the quinoprotein glucose dehydrogenase in the solubilization of exogenous phosphates by gram-negative bacteria. Phosphate Microorg. Molecular and Cellular Biology. ASM Press Wash. DC pp. pp. 197-203.
|
|
|
|
|
Gordon SA, Weber RP (1951). Colorimetric estimation of indoleacetic acid. Plant Physiology 26(1):192.
Crossref
|
|
|
|
|
Gouda S, Kerry RG, Das G, Paramithiotis S, Shin HS, Patra JK (2018). Revitalization of plant growth promoting rhizobacteria for sustainable development in agriculture. Microbiological Research 206:131-140.
Crossref
|
|
|
|
|
Gravel V, Antoun H, Tweddell RJ (2007). Growth stimulation and fruit yield improvement of greenhouse tomato plants by inoculation with Pseudomonas putida or Trichoderma atroviride: possible role of indole acetic acid (IAA). Soil Biology and Biochemistry 39:1968-1977.
Crossref
|
|
|
|
|
Gupta G, Parihar SS, Ahirwar NK, Snehi SK, Singh V (2015). Plant Growth Promoting Rhizobacteria (PGPR): Current and Future Prospects for Development of Sustainable Agriculture. Journal of Microbial and Biochemical Technology 7:96-102.
|
|
|
|
|
Gupta S, Meena MK, Datta S (2014). Isolation, characterization of plant growth promoting bacteria from the plant Chlorophytum borivilianum and in-vitro screening for activity of nitrogen fixation, phospthate solubilization and IAA production. International Journal of Current Microbiology and Applied Sciences 3:1082-1090.
|
|
|
|
|
Gyaneshwar P, Kumar GN, Parekh LJ, Poole PS (2002). Role of soil microorganisms in improving P nutrition of plants. Plant Soil 245:83-93.
Crossref
|
|
|
|
|
Herrmann L, Lesueur D (2013). Challenges of formulation and quality of biofertilizers for successful inoculation. Applied Microbiology and Biotechnology 97:8859-8873.
Crossref
|
|
|
|
|
Hillis DM, Dixon MT (1991). Ribosomal DNA: molecular evolution and phylogenetic inference. The Quarterly Review of Biology 66: 411-453.
Crossref
|
|
|
|
|
Hinsinger P (2001). Bioavailability of soil inorganic P in the rhizosphere as affected by root-induced chemical changes: a review. Plant Soil 237:173-195.
Crossref
|
|
|
|
|
Kloepper JW, Schroth MN (1978). Plant growth-promoting rhizobacteria on radishes. In Proceedings of the 4th International Conference on Plant Pathogenic Bacteria pp. 879-882.
|
|
|
|
|
Kloepper JW, Lifshitz R, Zablotowicz RM (1989). Free-living bacterial inocula for enhancing crop productivity. Trends in Biotechnology 7:39-44.
Crossref
|
|
|
|
|
Kumar A, Biotechnology D, Vishwavidyalaya GG (2016). Phosphate solubilizing bacteria in agriculture biotechnology: diversity, mechanism and their role in plant growth and crop yield. International Journal of Advanced Research 4:116-124.
Crossref
|
|
|
|
|
Lupwayi NZ, Clayton GW, Rice WA (2006). Rhizobial inoculants for legume crops. Journal of Crop Improvement 15:289-321.
Crossref
|
|
|
|
|
Marschner P, Crowley D, Yang CH (2004). Development of specific rhizosphere bacterial communities in relation to plant species, nutrition and soil type. Plant Soil 261:199-208.
Crossref
|
|
|
|
|
Milagres AM, Machuca A, Napoleao D (1999). Detection of siderophore production from several fungi and bacteria by a modification of chrome azurol S (CAS) agar plate assay. Journal of Microbiological Methods 37:1-6.
Crossref
|
|
|
|
|
Mirza MS, Ahmad W, Latif F, Haurat J, Bally R, Normand P, Malik KA (2001). Isolation, partial characterization, and the effect of plant growth-promoting bacteria (PGPB) on micro-propagated sugarcane in vitro. Plant Soil 237: 47-54.
Crossref
|
|
|
|
|
Olsen SR, Sommers LE (1982). Phosphorus. In: Page, A.L., Ed., Methods of Soil Analysis Part 2 Chemical and Microbiological Properties, American Society of Agronomy, Soil Science Society of America, Madison pp. 403-430.
|
|
|
|
|
Pereira SIA, Castro PML (2014). Phosphate-solubilizing rhizobacteria enhance Zea mays growth in agricultural P-deficient soils. Ecological Engineering 73:526-535.
Crossref
|
|
|
|
|
Pikovskaya R (1948). Mobilization of phosphorus in soil in connection with vital activity of some microbial species. Mikrobiologiya 17:362-370.
|
|
|
|
|
Reyes I, Valery A, Valduz Z (2007). Phosphate-solubilizing microorganisms isolated from rhizospheric and bulk soils of colonizer plants at an abandoned rock phosphate mine. In First International Meeting on Microbial Phosphate Solubilization, (Springer) pp. 69-75.
Crossref
|
|
|
|
|
Ribeiro CM, Cardoso EJBN (2012). Isolation, selection and characterization of root-associated growth promoting bacteria in Brazil Pine (Araucaria angustifolia). Microbiological Research 167:69-78.
Crossref
|
|
|
|
|
Rodriguez H, Fraga R (1999). Phosphate solubilizing bacteria and their role in plant growth promotion. Biotechnology Advances 17:319-339.
Crossref
|
|
|
|
|
Santoro MV, Cappellari L, Giordano W, Banchio E (2015). Systemic induction of secondary metabolite biosynthesis in medicinal aromatic plants mediated by rhizobacteria. In Plant-Growth-Promoting Rhizobacteria (PGPR) and Medicinal Plants, (Springer) pp. 263-285.
Crossref
|
|
|
|
|
Schwyn B, Neilands JB (1987). Universal chemical assay for the detection and determination of siderophores. Analytical Biochemistry 160:47-56.
Crossref
|
|
|
|
|
Shahab S, Ahmed N, Khan NS (2009). Indole acetic acid production and enhanced plant growth promotion by indigenous PSBs. African Journal of Agricultural Research 4:1312-1316.
|
|
|
|
|
Solanki M, Kundu BS, Nehra K (2018). Molecular diversity of phosphate solubilizing bacteria isolated from the rhizosphere of chickpea, mustard and wheat. Annals of Agrarian Science 16(4):458-463.
Crossref
|
|
|
|
|
Sureshbabu K, Amaresan N, Kumar K (2016). Amazing multiple function properties of plant growth promoting rhizobacteria in the rhizosphere soil. International Journal of Current Microbiology and Applied Sciences 5:661-683.
Crossref
|
|
|
|
|
Tsavkelova EA, Klimova SY, Cherdyntseva TA, Netrusov, A.I. (2006). Microbial producers of plant growth stimulators and their practical use: a review. Applied Biochemistry and Microbiology 42:117-126.
Crossref
|
|
|
|
|
Vessey JK (2003). Plant growth promoting rhizobacteria as biofertilizers. Plant Soil 255:571-586.
Crossref
|
|
|
|
|
Walpola BC, Arunakumara K (2016). Assessment of phosphate solubilization and indole acetic acid production in plant growth promoting bacteria isolated from green house soils of Gonju-Gun, South Korea. Tropical Agricultural Research and Extension 18 p.
Crossref
|
|
|
|
|
Weisburg WG, Barns SM, Pelletier DA, Lane DJ (1991). 16S ribosomal DNA amplification for phylogenetic study. Journal of Bacteriology 173:697-703.
Crossref
|
|
|
|
|
Wezel A, Rajot JL, Herbrig C (2000). Influence of shrubs on soil characteristics and their function in Sahelian agro-ecosystems in semi-arid Niger. Journal of Arid Environments 44:383-398.
Crossref
|
|