ABSTRACT
Curtoviruses are transmitted by the beet leafhopper Circulifer tenellus, in a circulative (non-propagative) manner. Curtoviruses are phloem-limited and are acquired by the vector during feeding. Sap-feeding insects harbor endosymbionts which can help provide essential nutrients required for the insects’ survival. Candidatus Sulcia muelleri is an endosymbiont present in the beet leafhopper identified during this study. A housekeeping gene, groel, was identified from the endosymbiont. The groel gene sequence from this strain of Ca. S. muelleri differs from all other strains published in NCBI, suggesting the presence of a new strain, which was named S. muelleri beet leafhopper (SMBLH). A GroEL-homolog protein produced from groel was found in different vectors with circulative transmission. Analysis of nucleotide and translated sequences, using alignment, phylogenetic trees, and predicted secondary and tertiary structures showed that SMBLH GroHp has similarities to Escherichia coli GroEL and the GroEL-homolog proteins from Hamiltonella and Buchnera, endosymbionts of whiteflies and aphids, respectively. GroHp and GroEL were expressed as fusion proteins. Electron microscopy analyses indicate that purified expressed GroHp and GroEL proteins demonstrate correct folding.
Key words: Beet leafhopper (BLH), Candidatus Sulcia muelleri, endosymionts, GroEL homolog protein (GroHp).
Plant viruses are economically important and can infect a wide range of host plants (Hogenhout et al., 2008). Insects play important roles in plant virus transmission. The insect vector acquires the virus while feeding on one plant then infects another plant when it takes its next meal (Hogenhout et al., 2008). Viruses must be able to survive, and sometimes replicate, in their insect hosts and still be transmissible and infective. Phloem feeding insects, such as mealybugs, aphids, and whiteflies, harbor bacterial endosymbionts. These endosymbionts may play roles important for the hosts’ survival. Wolbachia and Spiroplasma (endosymbionts of Drosophila flies) and Buchnera (aphids), provide protection against microbial pathogens (Shokal et al.,2016). Endosymbionts can provide the minimal host diet with essential nutrients (Douglas, 2009). Furthermore, they can also provide better temperature tolerance and parasite and insecticide resistance, as well as, sex determination (Montllor et al., 2002; Oliver et al., 2003; Kontsedalov et al., 2008).
Two types of bacterial endosymbionts can be found in insects, primary (obligate) and secondary (facultative) (Oliver et al., 2010). The location of the endosymbionts differs depending on the insect itself and the type of endosymbionts. Whiteflies that vector cotton leaf curl virus (CLCuV), harbor Portiera as their primary endosymbiont, and is found only in bacteriocytes, while the secondary endosymbiont Arsenophonus can be found in salivary glands, midgut, and bacteriocytes (Rana et al., 2012). For the aphid, Myzus persicae, the Buchnera aphidicila endosymbiont is restricted to bacteriocytes or mycetocytes found in the hemolymph (Bouvaine et al., 2011; van den Heuvel et al., 1994). Mealybug (Pseudococcidae, Hemiptera). Vectors of the ampelovirus grapevine leafroll-associated virus 3 (GLRaV-3) (Kono et al., 2008), have primary betaproteobacteria, endosymbiont (P-endosymbiont) that may act as a host for a secondary, gammaproteobacteria, endosymbiont (S-endosymbiont) (von Dohlen et al., 2001). It is very likely that insects acquire their primary endosymbionts through vertical maternal transmission (Baumann, 2005; Hogenhout et al., 1996), while secondary endosymbionts can be trans-mitted both vertically and horizontally (Oliver et al., 2010).
Leafhoppers belong to the order Hemiptera, suborder Auchenorrhyncha, family Cicadellidae (Moran et al., 2005). Auchenorrhyncha harbor the obligate endosymbiont Candidatus Sulcia muelleri. This bacterium is a nutritional endosymbiont belonging to phylum Bacteriodetes. It can be found in strap-shaped bacteriomes which can be found as a pair in the abdomen of adult insects (Moran et al., 2005; Moran, 2007). Although, different endosymbionts can be found in separate bacteriocytes in the insect body, Ca. S. muelleri along with Candidatus Baumannia cicadellinicola, a gammaproteobacterium, were found together in the same bacteriome of sharpshooters. In spittlebugs, Ca. S. muelleri was found together with Candidatus Zinderia insetticola, a betaproteobacterium (Wangkeeree et al., 2012; Moran, 2007). In the leafhopper Matsumuratettix hiroglyphicus, Ca. S. muelleri was associated with bacterium associated with M. hiroglyphicus (BAMH) in more than one region of single insect body, such as fat bodies, ovaries, and bacteriocytes (Wangkeeree et al., 2012). More than one type of facultative endosymbiont has been found in some leafhoppers but the different types of endosymbionts and the lack of regularity in finding them has prevented the formation of a clear account of the bacterial fauna in leafhoppers (Ishii et al., 2013).
Van den Heuvel et al. (1994) carried out research on the aphid, M. persicae, and a virus it transmits, potato leafroll virus (PLRV, Luteoviridae). The researcher discovered the presence of a protein produced by the aphid endosymbiont, Buchnera and named it symbionin. This protein can readily interact with the coat protein (CP) of PLRV. Analysis of symbionin, especially of the N-terminal, showed sequence homology with the Escherichia coli heat shock protein GroEL. Symbionin is a GroEL-like protein (GroHp) produced by Buchnera and is important for preserving the symbiont-aphid (Acyrthosiphon pisum) relationship (Ishikawa, 1982; van den Heuvel et al., 1997). GroHp is thought to interact and protect the virus while circulating in the insect, thus, facilitating its transmission (van den Heuvel et al., 1994). GroEL is a chaperonin, which reduces the number of aggregated proteins within the small confines of cells by assisting in the folding of proteins into their three dimensional structure (Skjaerven et al., 2015). The GroEL protein, or its homologues, can be found in all bacteria including endosymbiont bacteria. The function of GroHp in a virus/vector system could involve not only protecting the virus from degradation or from detection by the immune system, but also virus trafficking throughout the vector and preventing virus aggregation or disassembly (van den Heuvel et al., 1994; Morin et al., 1999; Gottlieb et al., 2010).
GroEL and GroHps are the most abundant proteins produced by bacteria (Baumann et al., 1996; Kupper et al., 2014). The groel gene is highly conserved in primary endosymbionts (Kupper et al., 2014).The beet leafhopper, Circulifer (Neoaliturus) tenellus (Baker) is a hemipteran insect (Cicadellidae) that transmits curtoviruses (Geminiviridae) which cause curly top disease. Curly top disease (CTD) is economically important affecting many plant crops including common bean, pepper, spinach, sugar beet, cucurbits, and tomatoes (Baliji et al., 2004). The endosymbiont(s) of the beet leafhopper (BLH) have not yet been identified. Furthermore, the GroHp produced by any endosymbiont(s) has not been explored. This report identifies the endosymbiont(s) of the BLH and analyzes a GroHp produced by the endosymbiont(s). The sequence of the groel gene responsible for this protein is analyzed. This gene has been amplified, cloned, and sequenced. The relationship between fourteen different GroHps produced by the endosymbionts of insect vectors has also been investigated and of C. tenellus was investigated and BLH GroHp predicted tertiary structure validity was determined using TEM imaging.
Leafhoppers and sugarbeets
The beet leafhoppers (C. (Neoaliturus) tenellus) used in this study were gifts from Carl Strausbaugh, USDA, Kimberly, ID, or collected from Las Cruces, NM, Leyendecker Plant Science Farm from Kochia scoparia plants.Beet leafhoppers were reared on sugarbeet plants maintained at 28°C day and 26°C night with a 16 h photoperiod. Adult insects were used for DNA extraction. The sugarbeet plants, Beta vulgaris, were grown from seeds of breeding line P1518-6 provided by Kelly Richardson, USDA, Salinas, CA.
DNA extraction from leafhoppers
Total DNA was extracted from whole beet leafhoppers using the “Purification of total DNA from insects” protocol of DNeasy Blood & Tissue Kit (QIAGEN Inc. Valencia, CA). The extracted DNA was diluted using molecular grade water to a final concentration of 30 ng/µl. It was stored at -20°C until needed.
Identification of C. tenellus endosymbiont(s)
To identify the symbiont(s) inhabiting C. tenellus, 16S rRNA primers (Table 1) were used (Noda et al., 2012). To test for specific endosymbionts known to colonize some phloem-feeding insects, specific PCR primers were used for Rickettsia (Noda et al., 2012), Wolbachia (Gonella et al., 2011), and Sulcia (Table 1). For the negative control, no DNA template was used in the amplification reaction. The PCR reaction contained 2.5 µl of 10X standard Taq reaction buffer (BioLabs, GA), 0.5 µl of 10 mM of dNTPs, 0.5 µl of 10 µM of each primer, 0.125 µl of Taq DNA polymerase (BioLabs, GA), 2 µl of DNA template, and nuclease-free water to a final volume of 25 µl.
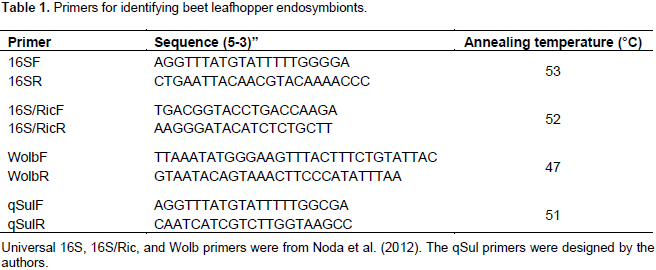
The PCR cycles were as follows: initial denaturation at 94°C for 5 min, 35 cycles of 94°C for 1 min, annealing cycle with various temperatures (Table 1), for 2 min, and 72°C for 1.5 min and a final extension at 72°C for 10 min, then ending at 4°C forever. PCR amplicons were electrophoresed in a 1% agarose gel in 1X TAE buffer (40 mM Tris-acetate and 1 mM EDTA), stained by GreenViewTM (GeneCopoeia, MD), and viewed under a UV light. The amplified products were cleaned using QIAquick PCR Purification kit (QIAGEN, MD), according to manufacturer’s instructions. The cleaned products were sequenced by MCLAB (South San Francisco, CA). Obtained sequences were compared with available sequences using NCBI Basic Local Alignment Search Tool (BLAST) algorithm.
Amplifying Ca. S. muelleri groel gene from C. tenellus
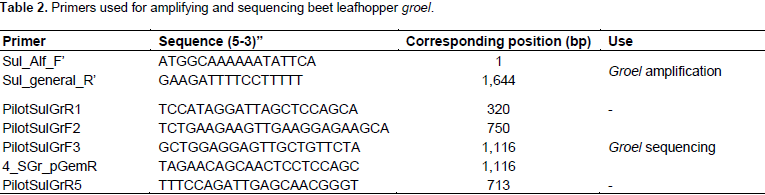
The Ca. S. muelleri groel gene was amplified from 2 µl of diluted
beet leafhopper DNA using primers designed from identical regions of
Ca. S. muelleri groel gene sequences of strains ALF, ML, DMIN, GWSS, BGSS, and PUNC (GeneBank accession no.
CP006060.1,
CP010105.1,
CP001981.1,
CP000770.2,
CP008986.1, and
CP013212.1, respectively). The PCR cycles were as follows: initial denaturation at 94°C for 5 min, 35 cycles of 94°C for 1.5 min, 42°C for 2 min, and 72°C for 1.0 min, and a final extension at 72°C for 10 min, then ending at 4°C forever. PCR amplicons were electrophoresed in a 1% agarose gel in 1X TAE buffer (40 mM Tris-acetate and 1 mM EDTA), stained by GreenView
TM (GeneCopoeia, MD). Table 2 lists all primers used for sequencing
groel.
Cloning of S. muelleri groel into pGEM-T Easy
Fresh (less than 24 h) groel PCR product was cleaned using QIAquick PCR Purification Kit (QIAGEN, MD) according to manufacturer’s instructions. The purified samples were eluted using molecular grade water and the concentration was measured using NanoPhotometerTM P-class spectrophotometer (IMPLEN, Germany). PCR amplicons of the groel gene were then cloned into pGEM-T Easy vector system (Promega, WI) according to instructions with a 5:1 ratio of insert to vector and incubation for 24 to 48 h at 4°C. The cloned plasmid was transformed into JM 109 high Efficiency Competent Cells (Promega, WI), also according to instructions.
The culture was plated on LB plates supplemented with 100 µl/ml ampicillin, 1 mM IPTG, and 20 mg/ml X-gal. The plates were incubated at 37°C for 12 h. White cells were tested using PCR colony method with groel amplification primers. PCR amplicons were electrophoresed using a 1% agarose gel in 1X TAE buffer (40 mM Tris-acetate and 1 mM EDTA), stained by GreenViewTM (GeneCopoeia, MD). Colonies containing inserts were inoculated on LB broth supplemented with ampicillin and incubated while shaking (200 rpm) at 37°C overnight. The plasmid was extracted using E.Z.N.A. Plasmid DNA Mini Kit I (OMEGA) and eluted using molecular grade water.
Sequencing of Ca. S. muelleri groel gene
The amplified and cloned Ca. S. muelleri groel gene-plasmid were cleaned using QIAquick PCR Purification Kit (QIAGEN, MD) and E.Z.N.A. Plasmid DNA Mini Kit I (OMEGA), respectively, according to manufacturers’ instructions. The concentrations of the cleaned products were measured using NanoPhotometerTM-P-Class. The concentration of amplified groel was adjusted to 20 ng/µl. The concentration of the purified groel clone plasmid was adjusted to100 ng/µl. Both samples were sequenced by MCLAB. Seven sequencing primers were used to sequence the complete groel gene (Table 2).
Analysis of sequences
The sequence of the complete
groel gene was assembled using Geneious (Biomatters Inc, Newark, NJ). This sequence was analyzed using NCBI BLASTN (
http://blast.ncbi.nlm.nih.gov/Blast.cgi) (Table 3). The
groel gene sequences of all
Ca. S. muelleri published in NCBI were aligned using Clustal Omega (
http://www.ebi.ac.uk/Tools/msa/clustalo/) and multiple alignments were created. The amino acid sequence of this
groel was obtained by translating the gene using EMBOSS Transeq (
http://www.ebi.ac.uk/Tools/st/emboss_transeq/). Multiple alignments of eight nucleotide and twelve amino acid sequences were generated using Clustal Omega. The sequences analyzed included all
groel genes of
Ca. S. muelleri strains published in NCBI, as well as,
E. coli, Hamiltonella, and
Buchnera groels. Phylogenetic trees of maximum likelihood were constructed using the program Phylogeny.fr (Dereeper et al., 2008, 2010), using “One Click” settings.
Beet leafhopper (BLH) endosymbiont,
Ca. S. muelleri GroEL-homolog protein (SMBLH GroHp) and the GroHps sequences of
E. coli, Hamiltonella, and
Buchnera, were used for structure prediction. Secondary structure was predicted using the PSIPRED Protein Sequence Analysis Workbench (
http://bioinf.cs.ucl.ac.uk/psipred/) (data not shown). The tertiary structure was predicted using SWISS-MODEL
https://swissmodel.expasy.org/interactive).
Cloning the genes
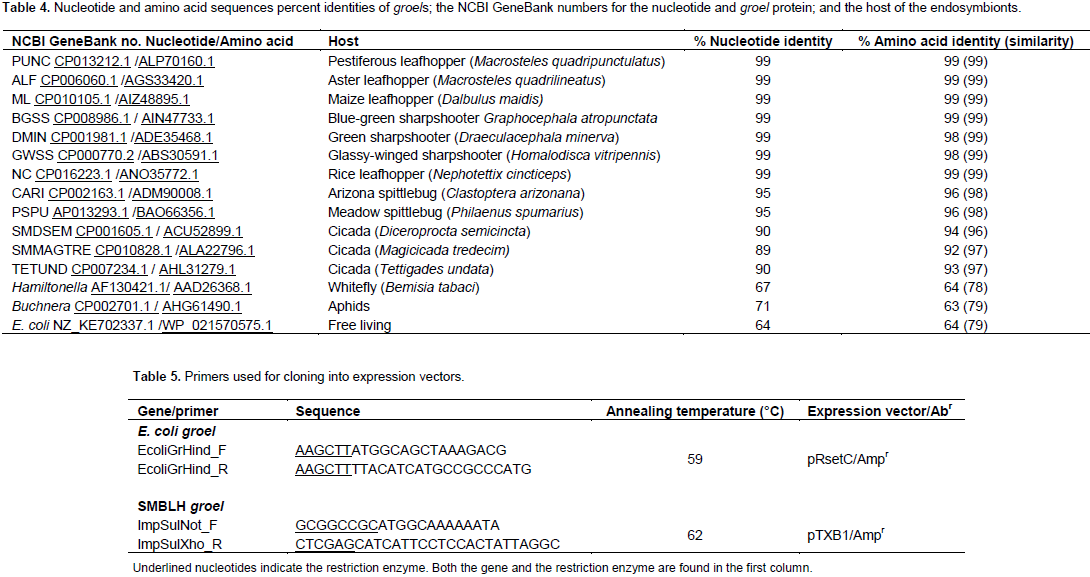
E. coli (MG6155) groel gene was cloned in the HindIII site (Table 5) of the expression plasmid pRsetC (Invitrogen, Carlsbad, CA). The resulting construct plasmid pRset C/EcoGroEL was propagated in E. coli BL21(DE3)lysS cells, on media supplemented with 100 µl/ml Amp and 30 µg/ml chloromphenicol. The SMBLH groel gene (without the stop codon), was cloned into NotI and XhoI sites (Table 5) of the expression vector pTXB1 (NEB, Ipswich, MA), upstream of the Intein site (IMPACT kitTM). The resulting construct plasmid pTXB1/SuGroHp was propagated in E. coli ER2566 cells, supplemented with 100 µl/ml Amp. The integrity of the genes was confirmed by sequencing.
Purification of over-expressed protein
Induction of pRset C and pTXB1 constructs with IPTG was performed according to instructions. Twenty five milliliters of LB supplemented with 100 μg/ml ampicillin was inoculated with a single colony of pLysS or E. coli ER2566 cells containing the construct plasmid, and incubated for 18 h at 37°C, while shaking at 220 rpm. Ten milliliters of culture was diluted in 1 L of LB supplemented with 100 μg/ml ampicillin, and incubated at 37°C while shaking until O.D600 = 0.4-0.6. Isopropylthio-β-Dgalactoside (IPTG) was added to a final concentration of 1 mM for pRset C clones and 0.4 mM for pTXB1 clones and the cells were incubated at 22°C for another 18 h. The cells were collected by centrifuging at 5,000 ×g for 15 min at 4°C. The supernatant was discarded and the pellet was kept at -80°C. The induction was assessed on a 10% SDS-PAGE and stained with Coomassie Brilliant Blue (CBB).
Purification of pRset C constructs
The induced protein pellet was resuspended on ice in 80 ml of 1X Native buffer (50 mM NaH2PO4, pH 8.0, and 500 mM NaCl, 10 mM imidazole, pH to 8.0). Then, sonicated five times, on ice at 60% amplitude for 30 s at 1 min intervals. The lysate was cleared by centrifugation at 15,000 ×g for 30 min at 4°C. The lysate was filtered through a 0.22 um filter then incubated with 10 ml of Ni-NTA beads (ThermoScientific, Waltham, MA) overnight on ice at 4°C, while rocking gently. The mixture was transported to a column and the flow through collected. The beads were washed four time with 100 ml of native buffer supplemented with 20 mM imidazole. The recombinant protein was eluted in 1 ml fractions (30 ml), with elution buffer (native buffer with 300 mM imidazole). The presence of protein in the fractions was tested on a 10% SDS-PAGE. The fractions with the protein were pooled, then concentrated using 30 kDa centricon.
Purification of pTXB1 constructs
The pellet was re-suspended on ice in cold 100 ml of column buffer (20 mM Tris-HCl, pH 8.5, 500 mM NaCl, 1 mM EDTA, 0.5% Triton-100, and 0.2% Tween 20), at 4°C. The re-suspended pellet was sonicated five times, on ice at 60% amplitude for 30 s at 1 min intervals. The lysate was cleared by centrifugation at 15,000 ×g for 30 min at 4°C. The lysate was filtered through a 0.22 µm filter and loaded to a 10 ml calibrated chitin column (NEB, Ipswich, MA). The chitin slurry was calibrated with 100 ml of column buffer. The lysate was allowed to flow through at a 0.5 to 1 ml/min. The chitin bed was washed with 200 ml of column buffer at a flow rate of 2 ml/min. On-column cleavage to release the protein was induced by a thiol reagent. 30 ml of cleavage buffer (20 mM Tris-HCl, pH 8.5, 500 mM NaCl, 1 mM EDTA, 50 mM DTT), was used to quickly flush the column. After the quick flush, the column was stopped leaving around 1.5 cm of cleavage buffer on top of the chitin bed. Incubate at 4°C for 24 to 30 h. The target protein was eluted from the column using 30 ml column buffer in a 1 ml fraction size. The presence of protein in the fractions was tested on a 10% SDS PAGE. The fractions with the protein were pooled then concentrated using 30 kDa centricon. The proteins were stored at 80°C.
Transmission electron microscopy (TEM)
Partially purified GroEL and GroHp were dialyzed in cold glycine/sodium hydroxide buffer, pH 8.1 (25 ml of 0.5 M glycine, titrated with 0.5 M NaOH). The samples were mounted on Carbon Film 200 mesh copper grids (Electron Microscopy Sciences, Hatfield, PA), then stained with 2.5% uranyl acetate. The samples were visualized in the bright field-imaging mode with a model H-7650 electron microscope (Hitachi High Technologies, Pleasanton, California, USA), at 50,000 ×magnification.
Detection and characterization of C. tenellus endosymbionts
The 16S rRNA gene is commonly used for identification of bacteria, because it is both conserved and ubiquitous, and targets a wide variety of bacteria. This gene was used to identify the endosymbionts of the beet leafhopper, C. tenellus, which vectors curtoviruses (Geminiviridae). The PCR amplification of the 16SrRNA gene gave a band of around 900 bp. The sequenced 16SrRNA genes were analyzed for similarity to sequences in the NCBI databank using the BLASTN algorithm and the top hits showed the endosymbionts to be mostly similar to proteobacterium (99% identity), acetobacteraceae (98% identity), and Asaia species (97% identity) (Table 3). Primers specific for the endosymbionts Ricketsia and Wolbachia were also tested but neither showed PCR products. Primers for Ca. S. muelleri (Table 1) amplified a product of size of around 210 bp. Sequence comparison using BLASTN algorithm when set for ‘Somewhat similar sequences’ identified the top hits of a 209 bp alignment with different strains of Ca. S. muelleri (99% identity), and a 199 bp alignment with Mycoplasma spp. (Table 3).
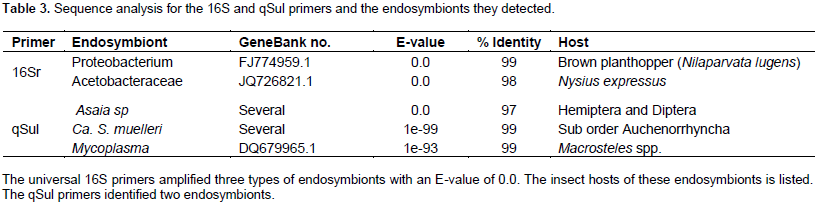
Ca. S. muelleri groel gene from C. tenellus
BLASTN and BLASTX were used to compare the groel from C. tenellus and 15 other groel sequences found in NBCI. Twelve of the 15 sequences belonged to different Ca. S. muelleri strains, while three were from Hamiltonella (whitefly endosymbiont), Buchnera (aphid endosymbiont), and E. coli (free living). Table 4 shows both nucleotide and amino acid sequences percent identities with the newly identified groel; the NCBI GeneBank accession numbers for the nucleotide and protein, and the host of the endosymbionts. Because this groel gene differs from all other groels from the strains of Ca. S. muelleri, it was deduced that it belongs to a new strain of Ca. S. muelleri, which can be found in BLHs.
The strain SMBLH (Sulcia muelleri beet leafhopper) was denoted. The sequence was submitted to GeneBank under the accession no. KY569409.The Ca. S. muelleri groel nucleotide sequence percent identities ranged from 90 to 99% compared to the beet leafhopper endosymbiont sequenced gene, while Hamiltonella, E. coli, and Buchnera, had 67, 64, and 71% identity, respectively. All Ca. S. muelleri groel amino acid sequences percent identities (and percent similarities), ranged from 92 (97) to 99 (99), compared to the beet leafhopper endosymbiont groel translated amino acid sequence, while Hamiltonella, E. coli, and Buchnera, had 64 (78), 64 (79), and 63% (79), identities, respectively.
Aligned Ca. S. muelleri groel sequences
All twelve Ca. S. muelleri groel sequences published in NCBI, and the newly sequenced (and translated) SMBLH were aligned using Clustal Omega. Figure 1 shows the first 60 nucleotides (the 5’-end) of groel gene of eight strains of Ca. S. muelleri (seven were published in NCBI and SMBLH). The strains TETUND, SMDSEM, SMMAGTRE, and PSPU, did not show enough identity at this region so they were omitted from comparison. At nucleotide 39, SMBLH was identical to the ML, ALF, PUNC, and NC strains. At the 3’-end, SMBLH lacked six nucleotides (GGTATG), when compared with DMIN, GWSS, BGSS, and (GGAATG) compared to ML, ALF, PUNC, and NC. But when comparing SMBLH to CARI, TETUND, SMDSEM, SMMAGTRE, and PSPU, lacked 15 nucleotides (not shown). DMIN, GWSS, BGSS, ML, ALF, PUNC, and NC have a 99% identity with SMBLH.
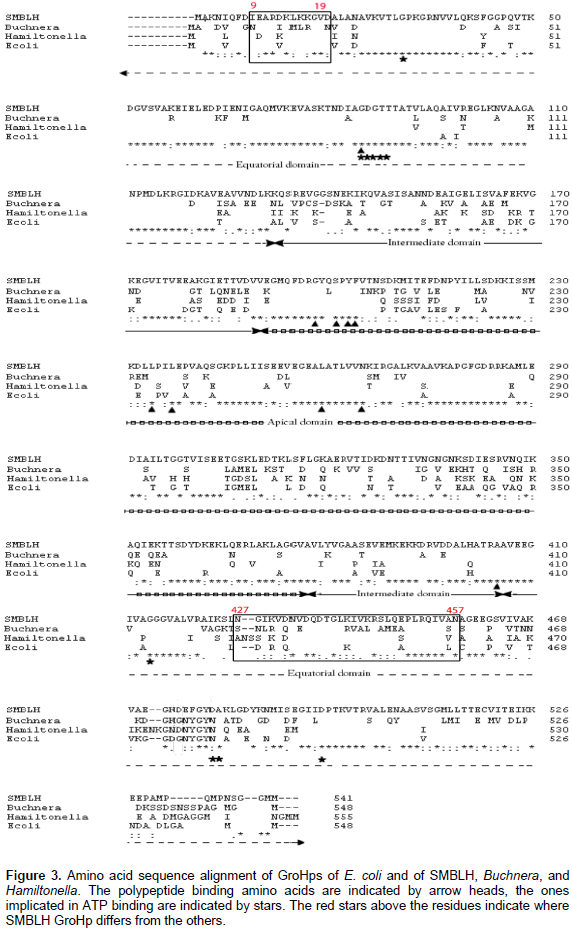
At the N-terminus SMBLH GroEL-homolog protein (GroHp) is identical to the first 50 amino acids of GroHps from Ca. S. muelleri BGSS, DMIN, GWSS, PUNC, ALF, and NC (Figure 2). At the C-terminus SMBLH GroHp lacks two amino acids (MG) at positions 536 and 537 when compared with CARI, BGSS, DMIN, GWSS, PUNC, ALF, and NC, while it lacks four amino acids, GG-MG (Figure 2). SMBLH has the same conserved amino acid residues as E. coli GroEL, with similarities to Buchnera GroHp, except for 474, where Asp is replaced with Gly, similar to Hamiltonella GroHp at this position. This substitution is at a conserved region. Furthermore, SMBLH shares the same polypeptide, and ATP binding sites as Buchnera, Hamiltonella, and E. coli, except for SMBLH and E. coli residues 479, where Asp is replaced with Asn. This change is at a less conserved region. Figure 3 shows the alignments and the polypeptide, and ATP binding sites, as well as the substituted residues.

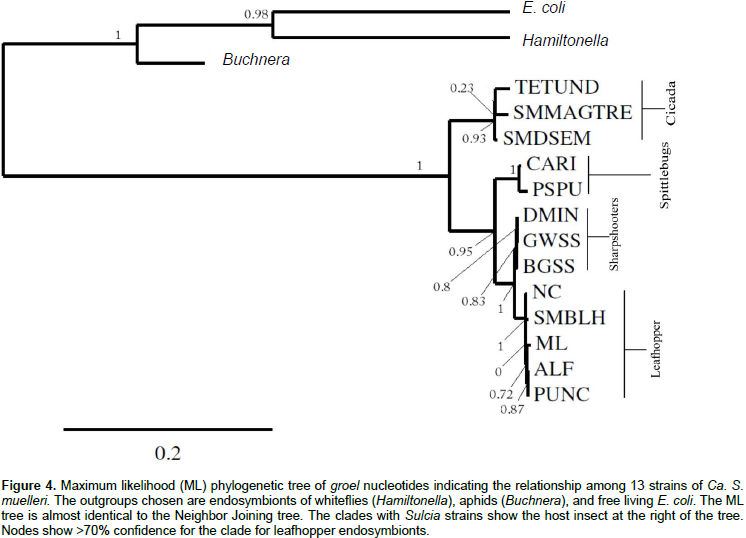
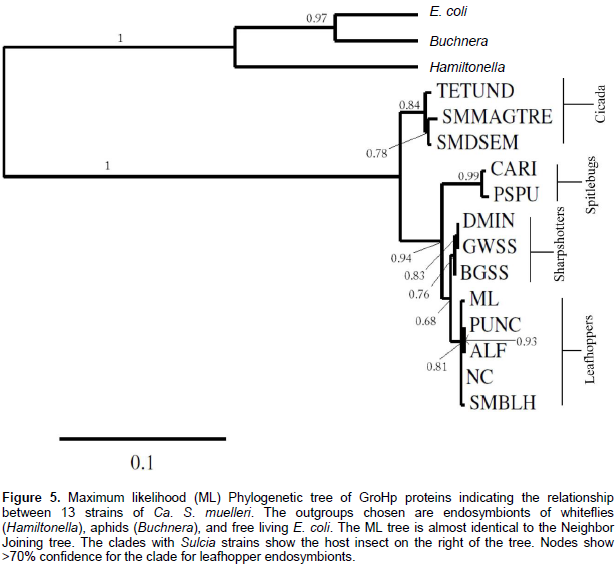
Phylogenetic analyses of groel and GroHp
Using maximum likelihood and neighbor joining, phylogenetic analyses were performed on 13 groels from different strains of Ca. S. muelleri, including SMBLH, for nucleotide and protein sequences, providing very similar results. The outgroups used were groels from Hamiltonella, Buchnera, and E. coli. The trees for nucleotide and amino acid sequences were highly concordant (Figures 4 and 5, respectively). All strains of Ca. S. muelleri were divided into two major clades with a strong support of 1 (Figures 4 and 5). The clade that SMBLH occupies has a support of 0.94 and greater. The phylogeny matches the grouping of endosymbionts and their host insect. The clades separate into cidada (TETUND, SMMAGTRE, and SMDSEM) and cicadellids (all others) (Figures 4 and 5 and Table 4). Furthermore, the clade of cicadellids divides into three branches, also in agreement with host, giving spittlebugs (CARI and PSPU), sharpshooters (DMIN, GWSS, and BGSS), and leafhoppers (SMBLH, NC, ML, ALF, and PUNC), the clade that has the SMBLH.
Prediction of the secondary and tertiary structures
The predicted secondary structures of SMBLH, E. coli, Hamiltonella, Buchnera, and GroHps show very similar motifs throughout the structures, except for three regions (data not shown). The residues between regions 184-191, 313-316, and 463-473, show similar and differences in motifs between the predicted secondary structures among the four GroHps (Table 6). In region 1 (residues 184-191), SMBLH GroHp has a β-strand motif, while the GroHps of E. coli, Buchnera and Hamiltonella, have a coil. In region 2 (residues 313-316), SMBLH GroHp had a coil motif, similar to that found in the GroHps of Buchnera and Hamiltonella, but unlike E. coli, which had an α-helix. At region 3 (residues 463-473), SMBLH GroHp had both β-strand and α-helix, while the GroHps from both E. coli and Buchnera had a β-strand and Hamiltonella had an α-helix.
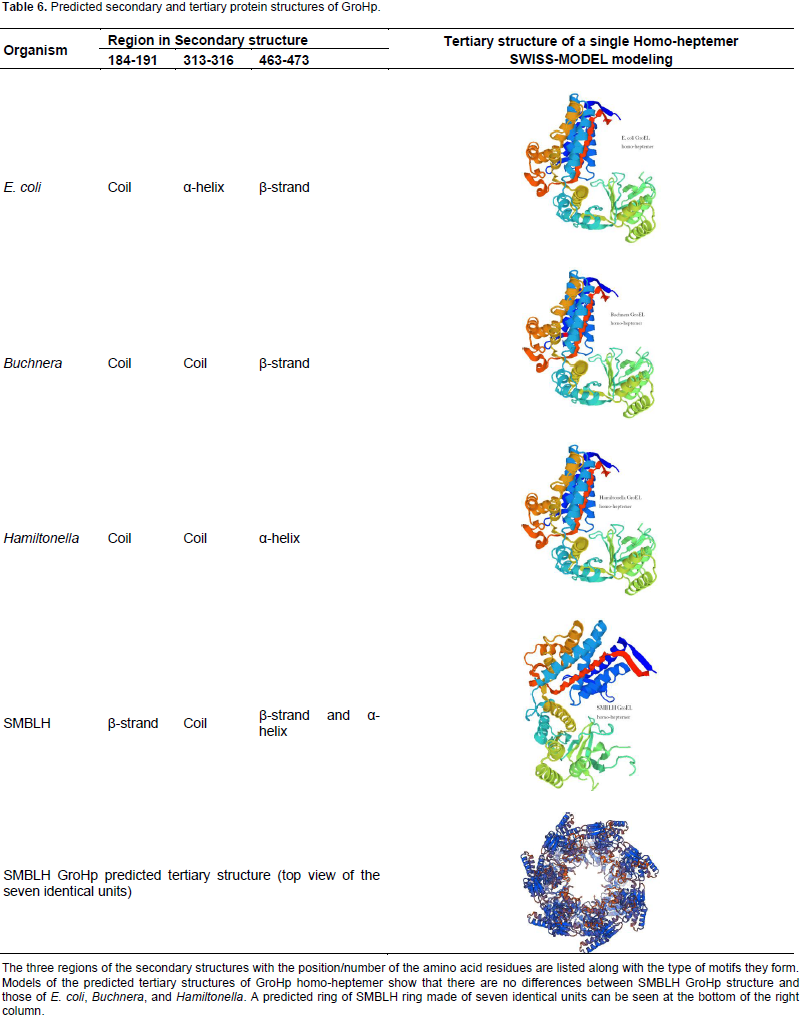
Table 6 shows the predicted homo-heptemer structure without a ligand. E. coli, Buchnera, and Hamiltonella GroHp proteins show similar tertiary structures. The GroHps from Ca. S. muelleri strains were represented by NC, TETUND, and SMBLH. The predicted structures of GroHps from Ca. S. muelleri strains NC, TETUND, and SMBLH were also similar to each other and to that of E. coli, Buchnera, and Hamiltonella predicted homo-heptemer structures, with mean model identities of 65. The predicted model for a 14-homomers for the SMBLH GroHp, showed it formed 2 rings, each with seven identical units, with a cavity in the middle. Table 6 shows the top view of the predicted SMBLH GroHp.
SMBLH GroHp TEM visualizing
Both partially purified GroEL and GroHp were visualized by TEM. GroEL structure appears to be folding as predicted (Figure 6). A ring of seven units was seen in the top view, while the side view displayed the staked rings. The SMBLH GroHp appeared to form similar structures, albeit the resolution was not as sharp as that of GroEL (Figure 6).
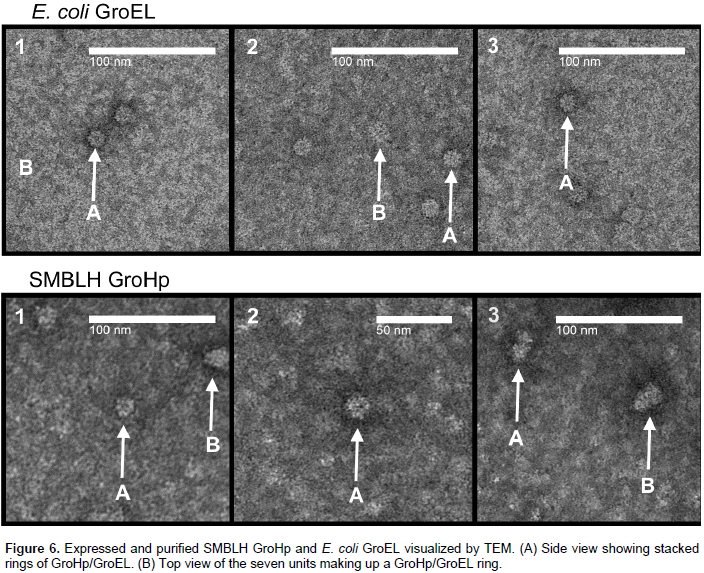
This study identified the endosymbiont Ca. S. muelleri from the beet leafhopper (BLH), C. tenellus. This was an expected result since Ca. S. muelleri is the primary endosymbiont of Auchenorrhyncha which includes the Cicadellidae family (leafhoppers). This endosymbiont has many strains, of which twelve complete genomes have been deposited into NCBI. This from all other groels produced by the Ca. S. muelleri published strains. It has the greatest identity to Ca. S. muelleri DMIN, GWSS, ML, ALF, PUNC, and NC. Because this groel was different to all other groels produced by any strain of Ca. S. muelleri published in NCBI, It was deduced that it belongs to a new strain not published in NCBI, which was named S. muelleri beet leafhopper (SMBLH).
The SMBLH groel length is shorter than the other strains groels by at least six bases at the 3’-end. The SMBLH GroEL-homolog protein (GroHp) sequence shows that it shares the same ATP binding sites and conserved regions as those of Buchnera, Hamiltonella, and E. coli GroHps. By aligning SMBLH GroHp with E. coli GroEL, it was found that both proteins retained the same conserved residues amongst these regions. The presence of different binding sites on the GroHp of endosymbiont Buchnera was explored by Hogenhout et al. (1998, 2000), by studying the binding of PLRV to GroHp. They found that the virus bound to the GroHp in the equatorial domain, mainly two regions.
One region contains residues 1 to 121 of the N-terminal and more specifically residues between 9 and 19. The other region contains residues 409 to 474 of the C-terminal, mainly residues between 427 and 457. In E. coli GroEL, the same domain exhibits polypeptide binding (Fenton et al., 1994). These regions in SMBLH GroHp still differ somewhat from Buchnera, Hamiltonella, and E. coli GroHp even in areas where they generally agree. The SMBLH GroHp differs at four residues between amino acids 9 to 19 and eight residues between 427 and 457. It is unknown if these differences play a role in binding to viruses, but the conservation of these regions is maintained.
Phylogenetic analyses using maximum likelihood and neighbor joining gave similar results. Both showed that SMBLH groel/GroHp had high identity with strains ML, ALF, PUNC, and NC of Ca. S. muelleri and all had leafhoppers as their host, such as Dalbulus, Macrosteles, and Nephotettix species. This is in agreement with Noda et al. (2012). The strains TESUND, SMMAGTRE, and SMDSEM with cicada as their host had the least homology with SMBLH groel/GroHp. The phylogeny was in agreement with their insect host suggesting co-evolution/co-speciation. Ca. S. muelleri is an endosymbiont of Auchenorrhyncha insects including leafhoppers, sharpshooters, spittlebugs, and cicada. This suggests that the strain SMBLH endosymbiont has the same evolutionary path and outcome as that of other Ca. S. muelleri strains.
Secondary structure was predicted for SMBLH GroHp, E. coli GroEL, Buchnera, and Hamiltonella GroHps. The structures show three regions with differences in the presence or absence of β-stand, α-helix, or coil motifs. SMBLH GroHp was different from the others in region 1 (at the intermediate domain) and region 3 (at the equatorial domain). Comparing these differences with residues at active sites, these differences do not appear to affect the binding sites or conserved regions. Tertiary structure prediction was employed to see if differences and similarities in the motifs of the predicted secondary structures would affect the folding. This folding might be more important for the protein function than the amino acid sequence itself. E. coli GroEL is made of two stacked rings. Each ring is made of seven identical subunits arranged together in a circle, forming 14 to homomer structure.
The predicted homo-heptemer structures for E. coli, Hamiltonella, Buchnera, and SMBLH, appeared very similar to each other. Furthermore, the predicted 14-homomer structure of SMBLH GroHp, is similar to the solved structure of E. coli GroEL (Braig et al., 1994). Thus, the differences in the secondary structure did not affect the tertiary folding. To validate the predicted tertiary structure of SMBLH GroHp, it was compared to that of E. coli GroEL. Both visualized E. coli GroEL and SMBLH GroHp, appeared to fold correctly. This further strengthens the conclusion that the differences in the motifs in the predicted secondary structure between the proteins did not have a role in folding.
Curly top disease (CTD) is economically important affecting many plant crops including common bean, pepper, spinach, sugar beet, cucurbits, and tomatoes (Baliji et al., 2004). Beet leafhopper (BLH) harbors a new strain of Ca. S. muelleri endosymbiont. It produces a GroHp that had not been previously identified. The role of aphids’ and whiteflies’ endosymbionts’ GroHp in plant virus transmission and plant resistance had been investigated. Interrupting the interaction between the virus’s coat protein (CP) and GroHp can reduce virus transmission capacity. This was done by feeding whiteflies, vector of begomoviruses (Geminiviridae), anti-GroHp antibodies derived from Buchnera GroHp, providing more than 80% reduction in virus transmission (Morin et al., 2000).
Even if the insect vector harbors more than one endosymbiont, only one GroHp derived from one of the endosymbionts has been implicated in virus transmission (Morin et al., 1999; Gottlieb et al., 2010; Rana et al., 2012; Su et al., 2013). GroHp could offer some resistance in transgenic plants which carry the gene for the whitefly endosymbiont GroHp protein. These plants were able to tolerate tomato yellow leaf virus (TYLCV), as well as Cucumber mosaic virus (CMV) infections. This was because both TYLCV and CMV were able to interact with GroHp, possibly trapping them in the plant and preventing movement of the virus (Edelbaum et al., 2009). Furthermore, GroHp from Xenorhabdus nematophila was used to bestow protection against the herbivorous insect Helicoverpa armigera, when ectopically produced by transgenic plants (Kumari et al., 2015).
The authors have not declared any conflict of interests.
REFERENCES
|
Baliji S, Black MC, French R, Stenger DC, Sunter G (2004) Spinach curly top virus: A newly described Curtovirus species from southwest Texas with incongruent gene phgylogenies. Phytopathology 94(7):772-779.
Crossref
|
|
|
|
Baumann P (2005). Biology of bacteriocyte-associated endosymbionts of plant sap-sucking insects. Ann. Rev. Microbiol. 59:155-189.
Crossref
|
|
|
|
|
Baumann P, Baumann L, Clark MA (1996). Levels of Buchnera aphidicola chaperoninGroEL during growth of the aphid Schizaphis graminum. Curr. Microbiol. 320:279-285.
Crossref
|
|
|
|
|
Bouvaine S, Boonham N, Douglas AE (2011). Interactions between a luteovirus and the GroEL chaperonin protein of the symbiotic bacterium Buchnera aphidicola of aphids. J. Gen. Virol. 92:1467-1474.
Crossref
|
|
|
|
|
Braig K, Otwinowski Z, Hegde R, Boisvert DC, Joachmiak A, Horwich AL, Sigler PB (1994) The crystal-structure of the bacterial chaperonin GroEL at 2.8 Angstrom. Nature. 371:578-586.
Crossref
|
|
|
|
|
Dereeper A, Audic S, Claverie JM, Blanc G (2010). BLAST-EXPLORER helps you building datasets for phylogenetic analysis. BMC Evol. Biol. 12:10-18.
Crossref
|
|
|
|
|
Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M, Claverie JM, Gascuel O (2008). Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36:465-469.
Crossref
|
|
|
|
|
Douglas AE (2009). The microbial dimension in insect nutritional ecology. Funct. Ecol. 23:38-47.
Crossref
|
|
|
|
|
Edelbaum D, Gorovits R, Sasaki S, Ikegami M, Czosnek H (2009). Expressing a whitefly GroEL protein in Nicotiana benthamiana plants confers tolerance to Tomato yellow leaf curl virus and Cucumber mosaic virus, but not to Grapevine virus A or Tobacco mosaicvirus. Arch. Virol. 154:399-407.
Crossref
|
|
|
|
|
Fenton WA, Kashi Y, Furtak K, and Horwich AL (1994). Residues in chaperonin GroEL required for polypeptide binding and release. Nature 371:614-619.
Crossref
|
|
|
|
|
Gonella E, Negri I, Marzorati, M, Mandrioli M, Sacchi, L, Pajoro M, Crotti E, Rizzi A, Clementi E, Tedeschi R, Bandi C, Alma A, Daffonchio D (2011). Bacterial endosymbiont localization in Hyalesthes obsoletus, the insect vector of Bois Noir in Vitis vinifera. Appl. Environ. Microbiol. 4(77):1423-1435.
Crossref
|
|
|
|
|
Gottlieb Y, Zchori-Fein E, Mozes-Daube N, Kontsedalov S, Skaljac M, Brumin M, Czosnek H, Vavre F, Fleury F, Ghanim M (2010). The transmission efficiency of Tomato yellow leaf curl virus by the whitefly Bemisia tabaci is correlated with the presence of a specific symbiotic bacterium species. J. Virol. 84 (18):9310-9317.
Crossref
|
|
|
|
|
Hogenhout SA, El-Desouky A, Whitefield AE, Redinbaugh MG (2008). Insect vector interactions with persistently transmitted viruses. Annu. Rev. Phytopathol. 46:327-359.
Crossref
|
|
|
|
|
Hogenhout SA, van der Wilk F, Verbeek M, Goldback RW, van den Heuvel JF JM (2000). Identifying the determinants in the equatorial domain of Buchnera GroEL implicated in binding Potato Leafroll Virus Potato leafroll virus. J. Virol. 74(10): 4541-4548.
Crossref
|
|
|
|
|
Hogenhout SA, van der Wilk F, Verbeek M, Goldback RW, van den Heuvel JF JM (1998). Potato leafroll virus binds to the equatorial domain of the aphid endosymbiotic GroEL homolog. J. Virol. 72(1):358-365.
|
|
|
|
|
Hogenhout SA, Verbeek M, Hans F, Houterman PM, Fortass N, van der Wilk F, Huttinga, H, van den Heuvel, JF JM (1996) Molecular bases of the interactions between luteoviruses and aphids. Agronomie. 16:167-173.
Crossref
|
|
|
|
|
Ishii Y, Matsuura Y, Kakizawa S, Nikoh N, Fukatsu T (2013). Diversity of Bacterial Endosymbionts Associated with Macrosteles Leafhoppers Vectoring Phytopathogenic Phytoplasmas. Appl. Environ. Microbiol. 79(16):5013-5022.
Crossref
|
|
|
|
|
Ishikawa H (1982) Host symbiont interactions in the protein-synthesis in the pea aphid, Acyrthosiphon pisum. Insect Biochem. 12(6): 613-622.
Crossref
|
|
|
|
|
Kono M, Koga R, Shimada M, Fukatsu T (2008). Infection Dynamics of Coexisting Beta- and Gammaproteobacteria in the Nested Endosymbiotic System of Mealybugs. Appl. Environ. Microbiol. 74(13):4175-4184.
Crossref
|
|
|
|
|
Kontsedalov S, Zchori-Fein E, Chiel E, Gottlieb Y, Inbar M, Ghanim M (2008). The presence of Rickettsia is associated with increased susceptibility of Bemisia tabaci (Homoptera: Aleyrodidae) to insecticides. Pest Manage. Sci. 64:789-792.
Crossref
|
|
|
|
|
Kumari P, Mahapatro GK, Banerjee N, Sarin NB (2015). Ectopic expression of GroEL from Xenorhabdus nematophila in tomato enhances resistance against Helicoverpa armigera and salt and thermal stress. Transgen. Res. 24:859-873.
Crossref
|
|
|
|
|
Kupper M, Gupta SK, Feldhaar H,Gross R (2014). Versatile roles of the chaperonin GroEL in microorganism–insect interactions. FEMS Microbiol. Lett. 353:1-10.
Crossref
|
|
|
|
|
Montllor CA, Maxmen A, Pircell AH (2002). Facultative bacterial endosymbionts benefits pea aphids Acyrthosiphon pisum under heat stress. Ecol. Entomol. 27:189-195.
Crossref
|
|
|
|
|
Moran NA (2007). Symbiosis as an adaptive process and source of phenotypic complexity. PNAS 104(suppl 1):8627-8633.
Crossref
|
|
|
|
|
Moran NA, Tran P, Gerardo NM (2005). Symbiosis and insect diversification: an ancient symbiont of Sap feeding insects from the bacterial phylum Bacteroidetes. Appl. Environ. Microbiol. 71(12):8802-8810.
Crossref
|
|
|
|
|
Morin S, Ghanim M, Sobol I, Czosnek, H (2000). The GroEL protein of the whitefly Bemisia tabaci interacts with the coat protein of transmissible and nontransmissible begomoviruses in the yeast-two-hybrid system. Virology 276:404-416.
Crossref
|
|
|
|
|
Morin S, Ghanim M, Zeidan M, Czosnek H, Verbeek M, van den Heuvel J (1999). A GroEL homologue from endosymbiotic bateria of the whitefly Bemisia tabaci is implicated in the circulative transmission of Tomato yellow leaf curl virus. Virology 256:75-84.
Crossref
|
|
|
|
|
Noda H, Watanabe K, Kawai S, Yukuhiro F, Miyoshi T, Tomizawa M, Koizumi Y, Nikoh N, Fukatsu T (2012) Bacteriome-associated endosymbionts of the green rice leafhopper Nephotettix cincticeps (Hemiptera: Cicadellidae). Appl. Entomol. Zool. 47:217-225.
Crossref
|
|
|
|
|
Oliver KM, Degnan PH, Burke GR, Moran NA (2010). Facultative symbionts in aphids and horizontal transfer of ecologically important traits. Annu. Rev. Entomol. 55:247-266.
Crossref
|
|
|
|
|
Oliver KM, Russell JA, Moran NA, Hunte MS (2003). Facultative bacterial symbionts in aphids confer resistance to parasitic wasps. PNAS 100(4):1803-1807.
Crossref
|
|
|
|
|
Rana VS, Singh ST, Priya NG, Kumar J, Rajagopal R (2012) Arsenophonus GroEL interacts with CLCuV and is localized in midgut and salivary glands of whitefly B. tabaci. PLoS ONE 7:1-13.
Crossref
|
|
|
|
|
Shokal U, Yadav S, Atri J, Accetta, Kenny E, Banks K, Katakam, Jaenike J, Elftherianos I (2016). Effects of co-occurring Wolbachia and Spiroplasma endosymbionts on the Drosophila immune response against insect pathogenic and non-pathogenic bacteria. BMC Microbiol. 16:1-16.
Crossref
|
|
|
|
|
Skjaerven L, Cuellar J, Martinez A, Valpuesta JM (2015). Dynamics, flexibility, and allostery in molecular chaperonins. FEBS Lett. 583:2522-2532.
Crossref
|
|
|
|
|
Su Q, Pan H, Liu B, Chu D, Xie W, Wang S, Xu B, Zhang Y (2013).Insect symbiont facilitates vector acquisition, retention, and transmission of plant virus. Nature Sci. Rep. 3(1367):1-6.
Crossref
|
|
|
|
|
van den Heuvel JF JM, Bruyere A, Hogenhout SA, Ziegler-Graff V, Brault V, Verbeek M, van der Wilk F, Richards K (1997) The N-terminal region of the luteovirus read through domain determines virus binding to Buchnera GroEL and is essential for virus persistence in the aphid. J. Virol. 71(10):7258-7265.
|
|
|
|
|
van den Heuvel JF JM, Verbeek M, van der Wilk F (1994). Endosymbiotic bacteria associated with circulative transmission of Potato leafroll virus by Myzus persicae. J. Gen. Virol. 75:2559-2565.
Crossref
|
|
|
|
|
von Dohlen CD, Kohler S, Alsop ST, McManus WR (2001). Mealybug β-proteobacterial endosymbionts contain γ-proteobacterial symbionts. Nature 412:433-436.
Crossref
|
|
|
|
|
Wangkeeree J, Thomas AM, Hanboonsonga Y (2012). Candidates for symbiotic control of sugarcane white leaf disease. Appl. Environ. Microbiol. 78(19):6804-6811.
Crossref
|
|