ABSTRACT
Antimicrobial surveillance and identifying the genetic basis of antimicrobial resistance provide important information to optimize patient care. The present study is an analytical cross sectional study aimed to determine the prevalence of multidrug resistant (MDR), extensively drug resistant (XDR), pan drug resistant (PDR) and extended-spectrum ?-lactamases genes among Gram-negative bacteria isolated in Saudi Arabia. A total number of 386 non-duplicate Gram-negative isolates were collected. Identification and susceptibility testing were done using automation system (BD Phoenix™). The extracted DNAs were subjected to multiplex polymerase chain reaction (PCR). The results showed that only 15 (3.9%) of isolates were fully susceptible, the overall prevalence of XDR, MDR, PDR was 129 (33.4%), 113 (29.3%) and 48(12.4%) respectively. High resistant rate was observed against the antibiotic agents of cephalosporins class 79.3% followed by the agents of penicillins class 69.4%. The most dominant resistant gene was bla SHV which was detected in 106/386 (27.5%) isolates, followed by bla CTX-M 90/386 (23.3%). Bla CTX-M showed significant relation with all used antibiotic except ampicillin/clavulanic acid, aztreonam, cefoxtin, and meropene. The isolates which showed frequent resistant genes were: Klebsiella pneumoniae 90/124 (72.6%), A. baumanni 37/67 (55.2%), and P. mirabilis 24/44 (54.5%). These findings underscore the need for optimization of current therapies and prevention of the spread of these organisms.
Key word: Multidrug resistant (MDR), extensively drug resistant (XDR), pan drug resistant (PDR), extended spectrum beta lactamase (ESBL), bla SHV, bla TEM, bla CTX-M.
Antimicrobial resistance is described as a condition in which the pathogens escape from the stress of the antibiotic exposure (Alam et al., 2017). The increasing incidence of antimicrobial resistance is a key concern globally and considered main obstacle in the treatment of patients suffering from bacterial infections (Patil et al., 2019). It has been estimated that about 1.3 to 2 fold rise in mortality is caused by antimicrobial resistant bacteria compared to susceptible infections (Alam et al., 2017). A dramatic evolution has occurred in the significance of infections caused by Gram-negative bacteria (GNB) and associated with considerable mortality (Alam et al., 2017; Patil et al., 2019; Paterson, 2008). The efficiency of the current prophylactic and empiric antibiotic treatment is compromised by the emergence of pan drug resistant (PDR), extensively drug resistant (XDR) and multidrug resistant (MDR) Gram-negative bacteria (GNB) (Patil et al., 2019; Paterson, 2008). The ability to escape from the antimicrobial effects may have contributed to the nature of these organisms, which are heterogeneous, complex group of plasmids-borne and rapidly evolving enzymes which are capable of hydrolyzing cephalosporins, penicillins, aztreonam and monobactams (Fernando et al., 2017; Provenzani et al., 2020).
The American society of infectious diseases identified six top priority dangerous pathogens producing extended spectrum β-lactamases (ESBLs). Three of these six pathogens are antibiotic resistant Gram-negative bacteria: Pseudomonas aeruginosa, Acinetobacter baumannii and Enterobacteriaceae (Lee et al., 2007). ESBLs have been classified into three major groups: bla SHV, bla CTX-M and bla TEM (Patil et al., 2019). Ting et al. (2013) stated that bla TEM, bla SHV and bla CTX-M genes are super-resistant extended spectrum b-lactamases. In 2017, the WHO published list of global priority pathogens, a catalog of twelve species of bacteria grouped according to their antibiotic resistance under three priority tiers: critical, high, and medium. The critical group involved three pathogens that were Gram-negative bacilli: Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacteriaceae (WHO, 2017). The WHO also notified that the level of resistance to antimicrobial drugs used to treat common infections is reaching a crisis point.
If world administrations do not control infections in order to slow down the growth of drug resistance, entire populations could be wiped out by superbugs (Perez et al., 2014; Ibrahim et al., 2010). There are few novel antibiotic classes targeting gram-negative bacterial infections in the pipeline (Provenzani et al., 2020). However the availability of regional information on the resistance rate is fundamental to implementing efficient treatment protocols against infectious pathogens and may help to prevent infections with multidrug resistance pathogens at the local level (Moolchandani et al., 2017; Ibrahim, 2018). Therefore, the present study is aimed to determine pattern of antimicrobial resistance and to detect ESBL genes among Gram-negative bacteria isolated in Tabuk city, Saudi Arabia.
The present study is an analytical cross sectional study, conducted in King Fahad Specialist Hospital and prince Fahad Bin Sultan Research Chair (University of Tabuk), Saudi Arabia. A total number of 386 non-duplicate Gram-negative isolates were collected in order to determine the prevalence of MDR, XDR, and PDR and to detect extended-spectrum b-lactamases genes bla SHV, bla CTX-M and bla TEM.
Identification and susceptibility test
Depending on the origin of the samples, each sample was cultured on suitable medium/ media from: MacConkey agar, CLED agar blood agar, and Chocolate agar or Brain Heart infusion broth. Then they were incubated aerobically at 37°C for 24 to 48 h except for the blood culture, which was incubated for 5 to 7 days in broth medium. Growths of corresponding organisms were further sub-cultured for purification purpose. The significant growth was identified to the species level. Identification and susceptibility testing were done using automation system (BD Phoenix™). Identified strains were tested in vitro against several antimicrobial classes including carboxypenicillin (Ticarcillin/Clavulanic acid), Penicillinase resistant penicillin (Ampicillin/Sulbactam and Piperacillin/Tazobactam), Cephalosporins (Ceftazidime and Cefepime), Aztreonam, Carbapenems (Ertapenem, Imipenem and Meropenem), Aminoglycoside (Amikacin, Gentamicin and Tobramycin), Fluoroquinolones (Ciprofloxacin and levofloxacin), Minocycline (Tetracyclin), Glycylcycline (Tigecycline), polymyxin E (Colistin), and sulpha drugs (Trimethoprim/Sulfamethoxazole). Antimicrobial selection for testing depends on types of isolates and site of samples which were done automatically by the program. Based on susceptibility test of the above mentioned antibiotic classes, the isolates were characterized as MDR, XDR and PDR.
Moreover thirteen agents of five antimicrobial classes which represented the most commonly used antibiotics, carboxypenicillin, penicillinase resistant penicillin, cephalosporins, aztreonam and carbapenems, were subjected for further study in order to determine the relation between these antibiotics and ESBL genes. These agents include: ampicillin, ampicillin/clavulanic acid, ticarcillin/clavulanic acid, aztreonam, piperacillin/tazobactam, cefalotin, cefoxitin, ceftazidime, ceftrixone, cefepime, imipenem, meropenem and ertapenem. Quality control and maintenance were achieved according to the manufacturer’s guidelines.
The BD Phoenix™ automated identification and susceptibility testing system empowers workflow efficiency, using automated nephelometry, which results in a standardized isolate inoculum and a reduction in potential technologist error along with accurate, reliable and rapid detection of known and emerging antimicrobial resistance (Carroll et al., 2006).
Detection of antimicrobial resistance genes
DNA was extracted from whole 386 GNB isolates, using boiling technique. Few colonies from each isolate were mixed with molecular biology-grade water (Eppendorf, Hamburg, Germany), the mixture were centrifuged at 15,000 × g for 5 min. The supernatant was discharged and the pellet was re-suspended in molecular biology grade water (Eppendorf, Hamburg, Germany) and subjected to boiling at 100°C in a water bath for 20 min, then cooled and centrifuged at 15,000 × g for 60s before it was stored at −20°C.
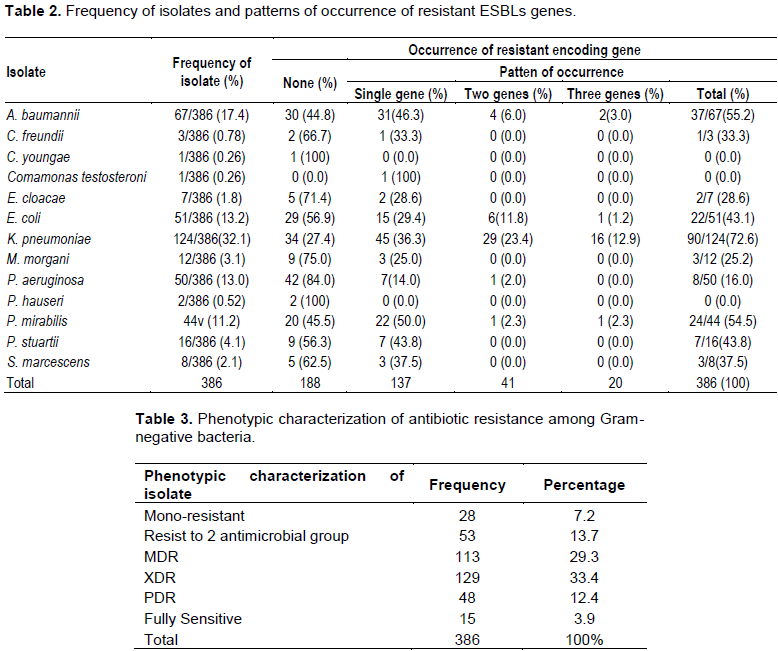
Multiplex polymerase chain reaction (PCR) was done to determine the presence of three ESBLs genes encoding bla TEM, bla SHV, bla CTX-M. Each extracted DNA was tested against the three sets of specific primers in a single test (Table 1). Amplification was performed in a final volume of 25µL containing 5mL of template DNA, 0.5 mL Taq polymerase, 1.0 mL of each primers and 0.2 mL dNTP mixture (10 mM), and finally the volume was completed to 25 µL by molecular biology-grade water to reach volume of 25 mL. The PCR was run according to the following protocol: essential denaturation at 95?C for 5 min followed by 40 cycle of denaturation at 95°C for 30s, annealing at 60°C for 30 sec and extension at 72°C for 1 min, the final step was extension at 72°C for 5 min. PCR product was run on 2% ethidium bromide agarose gel electrophoresis, and examined with gel imaging system, bands pattern was observed and interpreted according to their size (Table 1).
Analysis
The proportion of resistant for each antibiotic was calculated as the sum of resistant antibiotic relative to the sum of susceptible and resistant. The proportion of resistant class of antimicrobial represent the mean of resistance of all antimicrobial agents belong to that class. Chi-square tests were performed to determine the relation between ESBLs genes and antibiotic resistant using SPSS version 22. P value <0.05 was considered significant. The isolates which showed susceptibility to all groups of antimicrobial agents were classified as susceptible, while those that showed resistant to one group were classified as mono-resistant. The isolates resistant to one drug in two groups were classified as resistant to two antimicrobial groups. Multidrug resistant (MDR) denote when the isolates shown resistance to 3 or more antimicrobial group but susceptible to 2 or more. The US Centers for Disease Control and Prevention (CDC) and European Centre for Disease Prevention and Control (ECDC) have defined bacteria as pan drug resistant (PDR) when they are non-susceptible to all agents in all antimicrobial categories and as extensively drug-resistant (XDR) when they are non-susceptible to at least one agent in all, but two or fewer antimicrobial categories (Mohapatra et al. 2018). The ethical clearance for this study (UT-86-10-2019) was obtained from the research ethics committee, University of Tabuk (Saudi Arabia).
Table 2 shows the frequency of isolates along with patterns of occurrence of resistant ESBLs genes. The results revealed that the most common isolates were Klebsiella pneumoniae 124(32.1%), followed by A. baumanni 67(17.4%), E. coli 51(13.2%), P. aeruginosa 50(13.0%) and P. mirabilis 44(11.2%). The isolates showed high resistant rate against the antimicrobial agents of cephalosporins group, 79.3%, followed by the agents of penicillins, 69.4%; while against the agents of carbapenems they exhibited 32.5% resistance rate. In term of individual antimicrobial agent, the isolated Gram-negative bacteria revealed a high resistance rate against ampicillin 262(93.2%), followed by aztreonam 87(90.6%) and cefalotin 157(90.2%). Only 15(3.9%) of isolates were fully susceptible to all used antimicrobials. The overall prevalence of MDR, XDR, PDR was 113 (29.3%), 129 (33.4%) and 48(12.4%) respectively (Table 3).
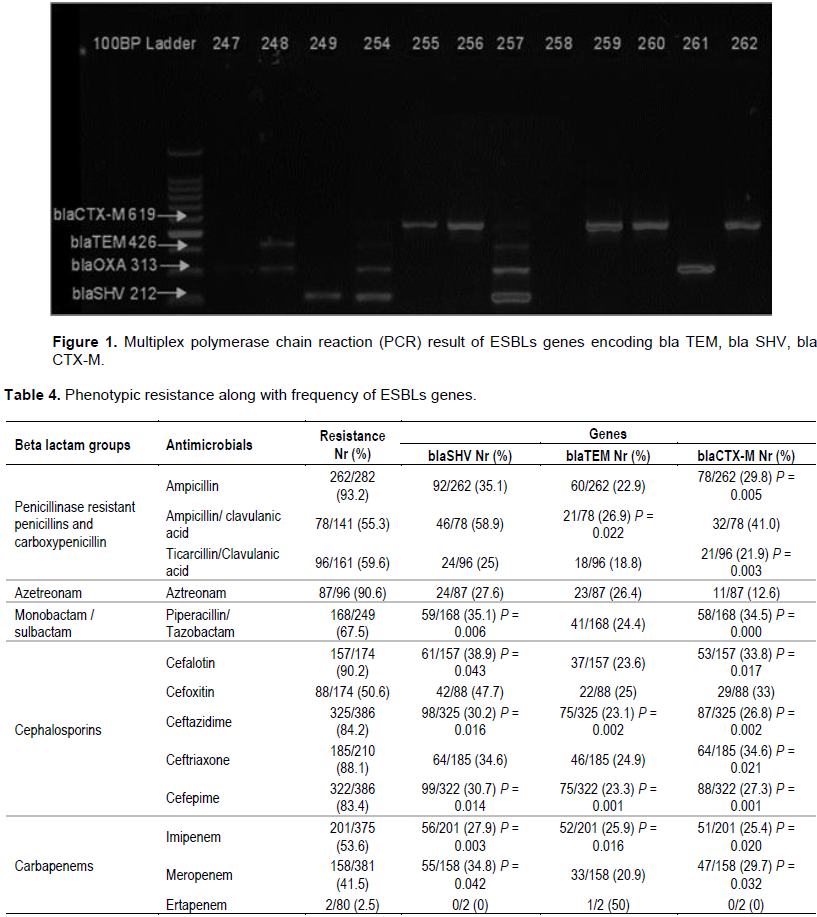
Screening for resistance genes showed that most of Gram-negative isolates harbored with resistance genes 198/386(51.3%), while isolates free from resistant genes were 188 (48.7%) (Table 2). Bla SHV was the most dominant gene, which was detected in 106/386 (27.5%) isolates; followed by bla CTX-M and bla TEM, which was detected in 90/386 (23.3%) and 78/386 (20.2%) isolates, respectively (Figure 1). Single resistant gene was detected in 137/386 (35.5%) isolates, coexistence of two genes were detected in 41/386 (10.6%) isolates, while triple genes were present in 20 (5.2%) isolates. Bla CTX-M showed significant relation with all used antibiotic except ampicillin/clavulanic acid, aztreonam and cefoxtin, while bla SHV exhibited significant statistic relationship with Piperacillin/ Tazobactam, Cefalotin, ceftazidime, cefepime, imipenem and meropenem. Bla TEM displayed significant relation only to ampicillin/clavulanic acid, ceftazidime, cefepime and imipenem. Ceftazidime and cefepime agents of cephalosporins class were least effective agents as they showed significant relation to the three resistant genes (Table 4). The isolates which showed the most frequent resistant genes were K. pneumonia 90/124 (72.6%), A. baumanni 37/67 (55.2%), E. coli 22/51 (43.1%), P. aeruginosa 8/42(19.0%) and P. mirabilis 24/44 (54.5%) (Table 2).
Bacterial resistant to different classes of antimicrobial agents are major threat to humanity and are of high risk, which may return the world to pre-antimicrobial era (Mohapatra et al., 2018; Koulenti et al., 2019). While active surveillance systems are set up in many countries in Europe, USA and Asia, little is reported on antimicrobial resistance status among Gram-negative bacteria in the Middle East, Africa and Saudi Arabia (Zowawi, 2016). The present study determined the prevalence of MDR, XDR, PDR and extended-spectrum- b-lactamases genes (TEM, SHV, CTX-M) among Gram-negative bacteria in Saudi Arabia.
The third generation cephalosporin such as ceftazidime and cefoperazone marked by stability to the common beta-lactamases of Gram-negative bacilli and these compounds are highly active against Enterobacteriaceae (Maina et al., 2012; Arumugham and Cascella, 2021). The isolated Gram-negative bacilli in this study showed high resistant rate to the antibiotic agents of cephalosporins class (79.3%), followed by the agents of penicillinase resistant penicillins which showed 69.4% resistant; while the agents of carbapenems had least resistant rate. The highest resistance rate was reported against ampicillin (93.2%), followed by aztreonam (90.6%) and cefalotin 157(90.2%). Similar trend of resistance was observed by Ruppé et al. (2015) who own the dramatic increase in the rates of resistance to third-generation cephalosporins to spread of plasmid-borne extended spectrum beta-lactamases (ESBLs) in Enterobacteriaceae and to occurrence of sequential chromosomal mutations, which may lead to the overproduction of intrinsic beta-lactamases, hyper-expression of efflux pumps, target modifications and permeability alterations in non-fermenting Gram-negative bacteria. The serious finding in our study was emerging of carbapenems resistant, Carbapenems agents considered the most active and potent agents against multidrug resistant (MDR) (Sheu et al., 2019). This finding is totally contradictory to that reported by Zaman et al. (2015) who determined the susceptibility pattern of Gram-negative bacilli isolated from a teaching hospital in Jeddah, Saudi Arabia and reported 100% sensitivity of enterobacteriaceae to carbapenems. Koulenti et al. (2019) concluded that carbapenem-resistant Enterobacteriaceae are particularly challenging to treat. However, WHO have listed carbapenem-resistant Enterobacteriaceae among the top tier of the antibiotic resistant that pose the greatest threat to human health (WHO, 2017).
The overall prevalence of MDR, XDR and PDR in this study was 29.3, 33.4, and 12.4% respectively. Several studies were conducted in Saudi Arabia and showed high resistance rate among Gram-negative bacteria, most of these studies focused on susceptibility per individual pathogen (Alam et al., 2017; Ibrahim et al., 2010; Ibrahim, 2018; Zowawi, 2016; Al Yousef, 2016; El-Saed et al., 2020). However, Ibrahim (2018), reported higher rate of MDR in Southwest Saudi Arabia (67.9%). The present study showed high PDR and less XDR compared to that reported by Mohapatra et al. (2018). Increasing antimicrobial resistant in Saudi Arabia may also be due to increased cross geographic transmission of drug resistant strains. Saudi Arabia is the capital of the Islamic world and has great number of expatriates, which makes it a potential center for the import and export of multi drug resistant strains (Ahmed?Abakur and Alnour, 2019), a recent study by Leangapichart et al. (2016) showed that returned travelers from Hajj had acquired MDR A. baumannii and NDM producing E. coli during the Hajj event.
The findings from this study showed that more than half (51.3%) of isolated Gram-negative harbored with resistant genes, while isolates free from resistant genes were 48.7%. Only 3.9% of isolates were fully susceptible to the used antimicrobials. This finding in alignment with Munita and Arias (2016) report and with Patil et al. (2019) results, who showed increase in the resistant rate among gram-negative pathogen and stated that Gram-negative bacteria are continuously evolving mechanisms to deactivate clinically important antimicrobial drugs by acquisition of resistance elements such as bla SHV, bla TEM and bla CTX-M. However antimicrobial resistant is an outcome of multifaceted microbial interactions such as microbial characteristics to gain resistance genes, selective pressure owing to inappropriate use and widespread of antibiotics, resistance may arise by the acquisition of de-novo mutation during treatment or by acquisition of integrative or replicative mobile genetic elements that have evolved over time in microbes in the natural ecosystem (Munita and Arias, 2016).
The results from this study indicate that bla SHV was the most prevalent resistant gene which was detected in (27.5%) isolates, followed by bla CTX-M (23.3%). Similar results indicated bla CTX-M and bla-SHV as the most prevalent genotypes of ESBLs producing gram-negative pathogens were reported in several countries (Paterson, 2008; Maina et al., 2012; Tian et al., 2010; Bajpai et al., 2017). Abrar et al. (2019) conducted three years study to determine the distribution of bla SHV, bla TEM, bla OXA and bla CTX-M genes in clinical isolates, they reported bla CTX-M as most dominant gene followed by bla OXA, bla TEM and bla SHV. Asokan et al. (2019) stated that the bla CTX-M gene indicates bacterial evolution due to cover prescription or weak enforcement of existing antibiotics policies. Moreover Provenzani et al. (2020) specified that the resistance rate grows rapidly when antibiotics are used inappropriately.
The present study showed that Klebsiella pneumoniae (72.6%), A.baumanni (55.2%), P.mirabilis (54.5%) E.coli (43.1%), and P.aeruginosa (19.0%) were the highest isolates harbored with of ESBLs genes. These findings are in agreement with Maina et al. (2012), Ibrahim (2018) and Asokan et al. (2019). Enterobacteriaceae such as K. pneumoniae, E. coli, Proteus spp, P. aeruginosa are naturally competent and can uptake naked DNA from the environment in suitable conditions (Patil et al., 2019). In our study bla CTX-M showed significant association to all used antibiotic agents of cephalosporin. This finding is in alignment with Maina et al. (2012), who stated that bla CTX-M type extended-spectrum β-lactamases (ESBLs), showing resistance to third and fourth-generation cephalosporins and to aztreonam. Similar result was reported by Nachimuthu et al. (2020).
Infections caused by multi-resistant Gram-negative pathogens negatively influence patient outcomes and costs. This study showed that only 3.9% of isolates had susceptibility to all used antibiotics, high resistant rate was observed against the antimicrobial agents of cephalosporins class and penicillinase resistant penicillin. The most dominant gene was bla SHV.
Infections caused by multi-resistant Gram-negative pathogens negatively influence patient outcomes and costs. This study showed that only 3.9% of isolates had susceptibility to all used antibiotics, high resistant rate was observed against the antimicrobial agents of cephalosporins class and penicillinase resistant penicillin. The most dominant gene was bla SHV.
The authors would like to thank the Deanship of Scientific Research (DSR) University of Tabuk, Saudi Arabia, for sponsoring the current research (project number S?1440?0339). They are very grateful for the valuable efforts put in during the sample collection by the Microbiology staff of King Fahad Specialist Hospital. They also appreciate Prince Fahad Bin Sultan Research Chair and Medical laboratory Technology staff, faculty of applied medical sciences, Tabuk University for their valuable support.
REFERENCES
|
Abrar S, Ain NU, Liaqat H, Hussain S, Rasheed F, Riaz S (2019). Distribution of blaCTX − M, blaTEM, blaSHV and blaOXA genes in Extended-spectrum-β-lactamase-producing Clinical isolates: A three-year multi-center study from Lahore, Pakistan. Antimicrobial Resistance and Infection Control 8:80.
Crossref
|
|
|
|
Ahmed Abakur EH, Alnour TM (2019). Detection of multidrug resistant Mycobacterium tuberculosis in Tabuk, Saudi Arabia, using genotype MTBDRplus. International Journal of Mycobacteriology 8:25 28.
Crossref
|
|
|
|
|
Alam MZ, Alam Q, Jiman-Fatani AA, shukri HA, Haque A (2017). A surveillance study on the prevalence and antimicrobial resistance pattern among different groups of bacteria isolated from Western province of Saudi Arabia. Biomedical Research India 28(2):898-906.
|
|
|
|
|
Al Yousef SA (2016). Surveillance of Antibiotic-Resistant Bacteria in King Khalid Hospital, Hafr Al-Batin, Saudi Arabia, During 2013. Jundishapur Journal of Microbiology 9(9):e19552.
Crossref
|
|
|
|
|
Arumugham VB, Cascella M (2021). Third Generation Cephalosporins.
View
|
|
|
|
|
Asokan GV, Ramadhan T, Ahmed E, Sanad H (2019). ITS Global Priority Pathogens List: A Bibliometric Analysis of MedlinePubMed for Knowledge Mobilization to Infection Prevention and Control Practices in Bahrain. Oman Medical Journal 34(3):184-193.
Crossref
|
|
|
|
|
Bajpai T, Pandey M, Varma M, Bhatambare GS (2017). Prevalence of TEM, SHV, and CTX-M Beta-Lactamase genes in the urinary isolates of a tertiary care hospital. Avicenna Journal of Medicine 7(1):12-16.
Crossref
|
|
|
|
|
Carroll KC, Glanz BD, Borek AP, Burger C, Bhally HS, Henciak S, Flayhart D (2006). Evaluation of the BD Phoenix automated microbiology system for identification and antimicrobial susceptibility testing of Enterobacteriaceae. Journal of Clinical Microbiology 44(10):3506-3509.
Crossref
|
|
|
|
|
El-Saed A, Balkhy HH, Alshamrani MM, Aljohani S, Alsaedi A, Al Nasser W, Gammal AE, Almohrij SA, Alyousef Z, Almunif S, Alzahrani M (2020). High contribution and impact of resistant gram negative pathogens causing surgical site infections at a multi-hospital healthcare system in Saudi Arabia. BMC Infectious Diseases 20:275.
Crossref
|
|
|
|
|
Fernando MMPSC, Luke WANV, Miththinda JKND, Wickramasinghe RDSS, Sebastiampillai BS, Gunathilake MPML, Silva FHDS, Premaratna R (2017). Extended spectrum beta lactamase producing organisms causing urinary tract infections in Sri Lanka and their antibiotic susceptibility pattern -A hospital based cross sectional study. BMC Infectious Diseases 17:138.
Crossref
|
|
|
|
|
Ibrahim ME (2018). High antimicrobial resistant rates among Gram-negative pathogens in intensive care units. A retrospective study at a tertiary care hospital in Southwest Saudi Arabia. Saudi Medical Journal 39(10):1035-1043.
Crossref
|
|
|
|
|
Ibrahim MK, Galal AM, Al-Turk IM, Al-Zhrany KD (2010) .Antibiotic resistance in Gram-negative pathogenic bacteria in hospitals' drain in Al-Madina Al-Munnawara. Journal of Taibah University for Science 3(1):14-22.
Crossref
|
|
|
|
|
Leangapichart T, Gautret P, Griffiths K, Belhouchat K, Memish Z, Raoult D, Rolain J (2016). Acquisition of a high diversity of bacteria during Hajj pilgrimage, including Acinetobacter baumannii with blaOXA-72, and Escherichia coli with blaNDM-5 carbapenemases. Antimicrobial Agents and Chemotherapy 60(10):5942-5948.
Crossref
|
|
|
|
|
Koulenti D, Song A, Ellingboe A, Abdul-Aziz MH, Harris P, Gavey E, Lipman J (2019). Infections by multidrug-resistant Gram-negative Bacteria: What's new in our arsenal and what's in the pipeline?. International Journal of Antimicrobial Agents 53(3):211-224.
Crossref
|
|
|
|
|
Lee JH, Jong SH, Lee SC (2007). A lack of drugs for antibioticresistant Gram-negative bacteria. Nature Reviews Drug Discovery 6:938.
Crossref
|
|
|
|
|
Maina D, Revathi G, Kariuki S, Ozwara H (2012). Genotypes and cephalosporin susceptibility in extended-spectrum beta-lactamase producing enterobacteriaceae in the community. Journal of Infection in Developing Countries 6(6):470-477.
Crossref
|
|
|
|
|
Mohapatra DP, Debata NK, Singh SK (2018). Extensively drug-resistant and pan drug-resistant Gram-negative bacteria in a tertiary-care hospital in Eastern India: A 4-year retrospective study. Journal of Global Antimicrobial Resistance 15:246-249.
Crossref
|
|
|
|
|
Moolchandani K, Sastry AS, Deepashree R, Sistla S, Harish BN, Mandal J (2017). Antimicrobial Resistance Surveillance among Intensive Care Units of a Tertiary Care Hospital in Southern India. Journal of Clinical and Diagnostic Research 11(2):DC01-DC07.
Crossref
|
|
|
|
|
Munita JM, Arias CA (2016). Mechanisms of antibiotic resistance. Microbiology Spectrum 4(2):1-37.
Crossref
|
|
|
|
|
Nachimuthu R, Kannan VR, Bozdogan B, Krishnakumar V, S KP, Manohar P (2020). CTX-M-type ESBL-mediated resistance to third-generation cephalosporins and conjugative transfer of resistance in Gram-negative bacteria isolated from hospitals in Tamil Nadu, India. Access Microbiology 3(3):000142.
Crossref
|
|
|
|
|
Paterson DL (2008). Impact of Antibiotic Resistance in Gram-Negative Bacilli on Empirical and De?nitive Antibiotic Therapy. Clinical Infectious Diseases 47(1):S14-S20.
Crossref
|
|
|
|
|
Patil S, Chen H, Zhang X, Lian M, Ren P, Wen F (2019). Antimicrobial Resistance and Resistance Determinant Insights into Multi-Drug Resistant Gram-Negative Bacteria Isolates from Paediatric Patients in China. Infection and Drug Resistance 22(12):3625-3634.
Crossref
|
|
|
|
|
Perez F, Adachi J, Bonomo RA (2014). Antibiotic-resistant gram-negative bacterial infections in patients with cancer. Clinical Infectious Diseases 15:59(Suppl 5):S335-S339.
Crossref
|
|
|
|
|
Provenzani A, Hospodar AR, Meyer AL, Leonardi Vinci D, Hwang EY, Butrus CM, Polidori P (2020). Multidrug-resistant gram-negative organisms: a review of recently approved antibiotics and novel pipeline agents. International Journal of Clinical Pharmacy 42:1016-1025.
Crossref
|
|
|
|
|
Ruppé E, Woerther PL, Barbier F (2015). Mechanisms of antimicrobial resistance in Gram-negative bacilli. Annals of Intensive Care 5:21.
Crossref
|
|
|
|
|
Sheu C, Chang Y, Lin S, Chen Y, Hsueh P (2019). Infections Caused by Carbapenem-Resistant Enterobacteriaceae: An Update on Therapeutic Options. Frontiers in Microbiology 10:80.
Crossref
|
|
|
|
|
Tian G, Garcia J, Adams-Haduch JM, Evangelista JP, Destura RV, Wang H, Doi Y (2010). CTX-M as the predominant extended-spectrum beta-lactamases among Enterobacteriaceae in Manila, Philippines. Journal of Antimicrobial Chemotherapy 65:584-586.
Crossref
|
|
|
|
|
Ting C, Jun A, Shun Z (2013). Detection of the common resistance genes in Gram-negative bacteria using gene chip technology. Indian Journal of Medical Microbiology 31(2):142-147.
Crossref
|
|
|
|
|
World Health Organization (WHO) (2017). Media Centre. News Release. WHO publishes list of bacteria for which new antibiotics are urgently needed. (2017).
View
|
|
|
|
|
Zaman R, Aly MM, Helmi NR (2015). Antimicrobial susceptibility pattern of Gram-negative bacilli isolated from a Teaching Hospital in Jeddah, Saudi Arabia. African Journal of Microbiology Research 9(41):2145-2158.
Crossref
|
|
|
|
|
Zowawi HM (2016). Antimicrobial resistance in Saudi Arabia An urgent call for an immediate action. Saudi Medical Journal 37(9):935-940.
Crossref
|
|