ABSTRACT
There is still unexplored reservoir of microorganisms from sediments and water within Lakes Olbolosat and Oloiden using culture dependent technique. The current study compares bacterial diversity within Lake Olbolosat a freshwater lake and Lake Oloiden a saline alkaline lake. Out of 60 isolates obtained from sediments and water samples, 35 were from Lake Olbolosat and 25 from Lake Oloiden. Microbial count ranged between 0-1.75× 105 cfu/ml from both lakes. There was a significant difference between bacterial density and sampling points (p<0.001, F=6.667), 58 were Gram-positive and 2 Gram-negative. Fifty-five isolates that were rod-shaped, 3 were cocci and 2 filamentous. There was excellent growth of isolates at an optimum growth pH range of 6-10, a temperature range of 25-30°C and a salinity range of 0-5%. There was significant difference at p<0.001 for bacterial growth within physiological parameters. The isolates utilized skimmed milk, starch, olive oil, cellulose powder and xylan, hence the production of extracellular enzymes. There was antimicrobial activity against Pseudomonas aeruginosa, Bacillus subtilis, Escherichia coli, Staphylococcus aureus and Candida albicans by bacterial isolates. BLAST analysis of partial sequences showed there were 4 different phyla. Firmicutes scored 77% closely affiliated with 20 strains, Actinobacteria scored 15% closely affiliated with 4 strains, Proteobacteria and Bacteroidetes scored 4% affiliated with 1 strain each. Novel bacteria from this study could provide insights into their diversity and biotechnological applications.
Keywords: Lakes, bacteria, sediment, water, culture-dependent.
Lake Olbolosat which means a marshy area in the Maasai language is a freshwater marine ecosystem and is endangered. The gradual drying up of this lake is probably due to human activities (Wafula and Murunga, 2020). Lake Oloiden, which means salty in the Maasai language, was also considered for its saline alkaline. The lake lacks water inflow resulting in saline-alkaline conditions (Maina et al., 2018). It is separated from its west shore by a peninsula. Studies in Kenya have so much focused on salty lakes like Lake Magadi, Lake Elementaita, Lake Nakuru and scanty on fresh and saline alkaline lakes. Microbes are ubiquitous (Laxma-Reddy et al., 2017). Saline alkaline and freshwater lakes are economically and ecologically important ecosystems due to their high productivity and nutrient recycling capacities. The input of nutrients and fast recycling is due to active anaerobes and aerobes microorganisms. Microbes are essential to the functioning and major biogeochemical cycles within lakes (Krivtsov et al., 2020). Culture-dependent technique is important for industrial application of the microbial isolates (Spini et al., 2018). Fresh and saline-alkaline lakes are ecosystems that can serve as models for studying microbial diversity. There is still unexplored reservoir of microorganisms from sediments and water within Lakes Olbolosat and Oloiden. Microbial communities have been mostly studied using culture-dependent techniques and due to the uncultivability of most microbes, very few organisms can be isolated from these lakes. However, culture-dependent technique helps in better understanding of microbial in physiology for industrial application (Spini et al., 2018). Culture-dependent technique cannot be used solely for the analysis of populations within microbial communities. Metagenomics, a culture independent technique, provides detailed information on the metabolic and functional capacity of a microbial community (Yadav et al., 2019). Culture-dependent technique studies from Lake Oloiden of the Kenyan Rift Valley revealed the presence of diverse populations of high G+C content belonging to the genus Artrobacter, Dietzia and Terrabacter (Duckworth et al., 1998). Bacteria of the genera, Pseudomonas Paenibacillus, Arthrobacter, Bacillus, Fictibacillus and Acinetobacter, have been isolated from L.Olbolosat (Wafula and Murunga, 2020). The current study is in line with the Kenyan government’s big four priority areas within the framework of vision 2030 whereby the department of Regional Development Management supports the conservation of natural resources through sustainable utilization and conservation of river basins and large water bodies (Kiunjuri, 2017).The study is in line with African Commission priority areas of Agenda 2063 framework on the use of indigenous knowledge in Science and technology and innovation for sustainable development that acts with a sense of urgency on climate change and environment (AU, 2015). Environmental conservation will contribute towards reaching the United Nations Millennium development goals to conserve marine resources for sustainable development, protect and restore the terrestrial ecosystem, hold and reverse land degradation, halt biodiversity loss and finally combat desertification (United Nations General Assembly, 2015). This study aims at isolating and characterizing novel bacteria from Lakes Olbolosat and Oloiden to provide insights into their diversity and biotechnological applications.
Research authorization
Research authorization was obtained from the National Commission for Science, Technology and Innovation (NACOSTI) (Research Permit Number NACOSTI/P/20/3808) and permission to obtain samples for research from Lakes Olbolosat and Oloiden (Reference Number KWS/BRM/5001) was obtained from the Kenya Wildlife Service (KWS).
Study site
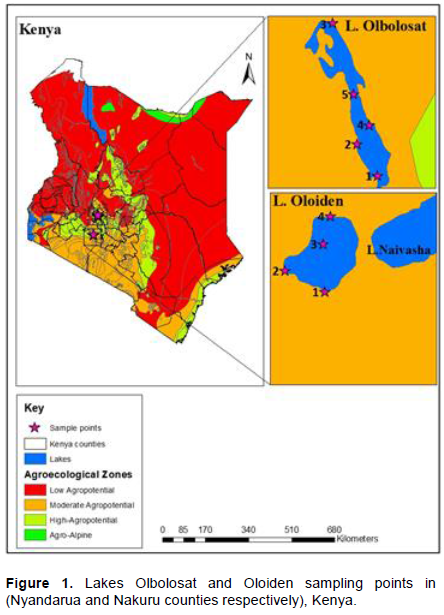
Lake Oloiden is about 4-7.5 km2. The lake is situated at a latitude of 0° 48'S and 36° 16'E (Maina et al., 2018). The lake lies at an average altitude of 1995 meters. The lake is a saline-alkaline lake that becomes fresh during rainy season caused by the overflow of Lake Naivasha (Maina et al., 2018). The lake records pH of 9 and temperature of 25°C. It is separated from its west shore by a peninsula. The distance between Lake Naivasha and Lake Oloiden are about 200 m. Lake Olbolosat is a freshwater body and is about 43 Km2 (Wafula and Murunga, 2020). The lake records pH of 7 and temp of 22°C. The lake is situated at a latitude of 0° 09'S and longitude of 36° 26'E in Nyandarua County in the central part of Kenya. It lies at an average altitude of about 2340 m in a wedge-shaped rift valley floor, known as Ongata Pusi Pusi, sloping in the eastward – northward direction. There is an underground inlet seeping under and emanates into the lake. From the Aberdare ranges, water from the basin flows northwards seeping to Thomson’s Falls into the northern part of Ewaso Nyiro River. It is formed by down warping and it is among the lakes in Kenya outside the rift valley (Figure 1).
Measurement of physico-chemical parameters
The geographical position of the sampling sites in terms of longitude, latitude and elevation were taken using Global Positioning System (GARMIN eTrex 20). The on-site metadata for temperature, electrical conductivity (EC), total dissolved solids (TDS) and dissolved oxygen (DO) of each sampling point were measured using Electrical Chemical Analyzer (Jenway - 3405) The pH was measured with a portable pH-meter (Oakton pH 110, Eutech Instruments Pty. Ltd) and confirmed with indicator strips (Merck, range 5-10) (Table 1).
Sample collection
Nine sampling points were selected randomly. Four from Lake Oloiden and 5 from Lake Olbolosat. Wet, dry sediments and water samples were randomly collected in triplicates. The experimental design used in the current study was purposive. The sample size was determined based on the unique features of the optimum coverage. There were three biological replicates for all water, wet and dry sediments. This was done by scooping wet and dry sediments with a hand shovel into sterile 250 ml plastic containers. The sterile plastic containers were used to fetch water from both lakes. All samples were transported on dry ice to the laboratory at Jomo Kenyatta University of Agriculture and Technology (Table 1).
Isolation and enumeration of bacterial isolate
Ten milliliter of water was suspended in 90ml of ringer salt solution powder (RSSP Himedia-M525) consisting of Sodium Chloride 8.50 g, Potassium Chloride 0.20 g, Calcium chloride anhydrous 0.20 g and Sodium bicarbonate 0.01g. Ten grams of the dry and wet sediments were also suspended separately in 90 ml of ringer solution. This was followed by filtration through sterile 125mm (Whatman ®) qualitative filter paper, Grade 1(Merck). One ml of the filtrate was transferred to 90ml of ringer solution to make 10-2 and10-3. The inoculation mixture with serial dilution was then spread in triplicate on the plates containing Plate count agar (PCA) Himedia- M091S) for the bacterial diversity. The medium consisted of Casein enzymic hydrolysate 5 g, Yeast extract 2.50 g, Dextrose 1.00 g and agar 15 g in 1 litre of water from the lakes to mimic the lake conditions. This was followed by incubation at 30°C for 24 to 72 h. To measure survival efficiency, colonies were counted using the following formula (Das and Dutta, 2018).
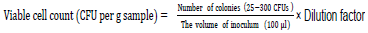
To obtain pure cultures, distinctive colonies were picked, transferred to fresh media and incubated at 30°C for 24 to 72 h. Purified colonies were grown on nutrient broth (Difco) and stored in 20% glycerol at -75°C.
Morphological and cultural characterization of bacterial isolates
Morphological and cultural characterization of the isolates was done under the dissecting and compound×100 microscope to observe pigmentation, form, elevation, margin, cell size, shape and arrangements as described by Cappuccino and Sherman, 2014). Classical Gram-staining and catalase tests were performed and the Gram-reaction confirmed by 3% (w/v) KOH test according to Moyes et al. (2009).
Physiological and biochemical characterization of bacterial isolates
The isolates were determined for their ability to grow at different temperatures, pH and also at different salt concentrations using PCA medium. The ability of the isolates to grow at different temperatures ranges were determined by growing the isolates at 25, 30, 35, 40, 45 and 50°C. The ability of isolates to grow at different pH ranges was determined by growing the isolates at 30°C at pH4, pH6, pH8, pH10, pH12 and pH14 adjusting each pH using 1M of HCL or NaOH. Salt tolerance was also determined by growing the cultures at 30°C with the media supplemented with 0, 5, 10, 15, 20, 25 and 30% NaCl concentrations (Vinet and Zhedanov, 2010). Intracellular and extracellular enzyme activities were determined according to (Cappuccino and Sherman, 2014). The biochemical tests included sugar utilization, catalase, urease, gelatin liquefaction, motility, starch and IMVIC.
Screening of bacterial isolates for hydrolytic enzyme production
The bacterial isolates were screened qualitatively for the production of five important enzymes such as xylanase, amylase, lipase, cellulase and protease. The bacterial isolates were cultured separately on the substrates such as xylan, starch, olive oil, cellulose powder and skimmed milk amended agar plates respectively (Mohammad et al., 2017). The isolates were then incubated at 30°C for 24-48 h. After growth Petri dishes were flooded with indicator solution. The negative control consisted of the uninoculated plate.
Screening the bacterial isolates that produce antimicrobial activity
Sixty bacterial isolates were screened for their ability to inhibit the growth of bacterial test organisms; Pseudomonas aeruginosa (ATCC 27853), Escherichia coli (NCTC 10418), Staphylococcus aureus (NCTC 10788), Bacillus subtilis (ATCC 55732) and fungal test organism Candida albicans (ATCC 90028) obtained from Kenya Medical Research Institute- Centre for Microbiology research. The bacterial isolates were cultured in nutrient broth and incubated at 30°C for 24 h. The cultured bacterial isolates were centrifuged at 10,000x g for 1min and the supernatant sieved using sterile micro membrane filters to remove any bacterial cells. The impregnated sterile Whatman ® qualitative filter papers, Grade 1(Merck) discs measuring 1centimeter paper discs were aseptically placed on Mueller Hinton agar (Himedia-M173). The media was swabbed with 0.1 ml per Petri dish of the test organisms following Kirby-Bauer diffusion protocol followed by incubation for 24-48 h at 30°C after which the results were recorded while negative control consisted of the uninoculated plate (Hudzicki, 2012).
Molecular characterization of bacterial isolates using partial 16S rRNA genes
Bacterial isolates were grown in nutrient broth media (Himedia-M002) consisting of Peptone, 5g, sodium chloride, 5 g, HM peptone B# 1.5 g, Yeast extract, 1.5 g in 1 L of distilled water. The overnight cultures were centrifuged at 10,000× g for 1 min and the supernatant discarded remaining with the pellet. DNA was extracted using QIAamp DNA Mini Kit (Qiagen, Germany) extraction kit according to manufacturer’s instructions. Bacterial universal primers 27F forward (5′-AGA GTT TGA TCC TGG CTC AG-3′) and 1492R reverse (5′-GGT TAC CTT GTT ACG ACT T-3′) were used for the amplification of 16S rRNA gene. PCR was carried out using PEQLAB, Erlangen, Germany, 96 PCR thermocycler machine. The PCR was carried out in a 50 μl mixture containing 25 μl 3X Taq PCR Master Mix (Qiagen, Germany), 2.5 μl of each primer, 10 μl of DNA template (50 ng) and 10 μl RNase free water. The reaction mixtures were subjected to the following PCR conditions: Initial activation of the enzyme at 95°C for 5 min followed by 32 cycles consisting of 1-min denaturation at 95°C for 1-min, primer annealing at 55°C for 2 min, chain extension at 72°C for 1.5 min and a final extension at 72°C for 10 min (Roux, 1995). The amplified PCR products were checked by gel electrophoresis using 1.2% (w/v) agarose gels stained with ethidium bromide (1 μg/ml) and visualized using Biotec-Fischer Felix6050 gel documentation system (ProfiLab24, Germany) according to the manufacturer’s instructions and stored at -20°C. Purification and Sanger sequencing of PCR products of the 60 bacterial isolates were carried out at Human Genomics Macrogen Europe (Macrogen Europe B.V, Amsterdam, Netherlands).
Statistical analysis
Data on bacterial density was noted and recorded in an Excel sheet. Two Way Analysis of Variance was used to analyze all measured data. Normality Test (Shapiro- Wilk) was used to compare means using Sigma Plot 12 v 5.0 for bacterial density. Correlation profiles of zones of hydrolysis and bacterial isolates zone of clearance were visualized as heatmaps generated by a hierarchical clustering R script using R v 4.0.2. Sequencing was conducted in one direction using the forward primer (27 F). The Chromas pro program was used to remove ambiguity and comparisons were done with the NCBI GenBank databases using Basic Local Alignment Search Tool (BLAST) and EZBio Cloud algorithms. Sequences were submitted to the GenBank database and were assigned the accession numbers. The differences in the nucleotides were converted into distance matrices using the Maximum Likelihood method (Saitou and Nei, 1987). A phylogenetic tree was constructed using MEGA 7 (Engeset et al., 2003).
Sampling
Sediments and water samples were randomly collected in triplicates. The metadata collected before sampling included the geographical positions of each site in terms of latitude, longitude, elevation, temperature, pH, electrical conductivity, total dissolved solids and dissolved oxygen. Samples were collected from the two lakes and parameters summarized in Table 1.
Isolation of bacterial isolates obtained from Lakes Olbolosat and Oloiden
A total of 60 bacterial isolates were obtained from lakes Olbolosat and Oloiden. Lake Olbolosat recorded 35 while Lake Oloiden recorded 25 bacterial isolates (Table 2).
Enumeration of bacterial isolates obtained from Lakes Olbolosat and Oloiden
The microbial counts for dry sediments ranged between 0 to 1.75 × 105 cfu/ml respectively. The microbial counts for wet sediments ranged between 7.63×104 to1.16 ×105 cfu/ml respectively while the microbial count for water samples ranged between 5.3×104 to 1.22×105 cfu/ml. Bacterial density significantly varied (p<0.001, F=6.667) between the sampling points (Figure 2 and Plate 1a and b).
Cultural characterization
The isolates had varying colony characteristics. Most bacterial isolates were cream white, some were white, and a few were cream; while the rest were cream yellow, orange, watery, reddish and brown. The highest percentage of isolates were circular, a few were irregular, filamentous and only one was concentric in form. The margin of most isolates was entire while a few were undulate, serrated or lobate. The highest percentage of the isolates had a flat elevation while others were umbonate, pulvinate, or raised (Table 2 and Plate 2a -d). A dendrogram for morphology and cellular characteristics based on Ward D method and the distance between characters measured using Euclidean metric for the hierarchical clustering (Figure 3).
Cellular characterization
Cellular characterization revealed two isolates that were gram-negative out of sixty isolates. Fifty-seven isolates were rod-shaped, two were cocci and one was filamentous in shape (Table 2 and Plate 3a-d). A dendrogram for morphology and cellular characteristics based on Ward D method and the distance between characters measured using Euclidean metric for the hierarchical clustering (Figure 3).
Biochemical characteristics of bacterial isolates
Biochemical tests for the bacterial isolates recorded the following positive results for intracellular and extracellular enzyme activities; all the 60 isolates were positive for catalase, twenty-six for citrase. forty-six for Methyl Red, seventeen for Voges-Proskauer, ten for urea, twelve for indole, twenty-nine for gelatin liquefaction, thirty-seven for motility, and forty-four for starch. Triple Sugar Iron (TSI) test had twenty-four that were positive for the production of acid and twenty-one for the production of alkaline and seven for hydrogen gas production (Table 3).
Physiological characterization
The isolates were able to grow at a wide range of pH including acidic, neutral and alkaline. There was poor growth at pH 4 and 12, while there was good growth at pH 6, 8 and 10. Salinity that favored the growth was 0% followed by 5%. The best temp for the microbes was at 30° while 25° followed. There was significant difference at p<0.001 for bacterial growth in all parameters; Kruskal-Wallis One Way Analysis of Variance on Ranks followed by Tukey’s HSD post-hoc analysis (Figure 4).
Hydrolase activity
The ability of bacterial isolates to produce extracellular enzymes was studied. Clustering from the heatmap shows that most bacterial isolates were able to utilize different substrates indicating their ability to produce different enzymes. Correlation between enzyme hydrolysis activity and bacterial isolates revealed that there were two functional clusters. Fifty-nine isolates formed a single cluster while ECP 3.1 formed a solitary cluster. Among the five enzymes assayed, all others formed a common cluster while xylanase formed a solitary cluster. This shows that some of the isolates were not able to utilize xylan as a substrate. Hydrolase activity that recorded positive result was indicated by the clear zone around the colony (Plate 4a-d and Figure 5).
Antimicrobial activity
Antagonistic activity recorded positive results that were indicated by a zone of inhibition around the colony. Zone sizes for inhibitions were looked up on a standardized chart by following Kirby-Bauer diffusion protocol for the sensitivity measuring above 18 mm, resistant 13 mm or less and intermediate measuring 14-16 mm. Clustering from the heatmap shows that most bacterial isolates were sensitive (measuring >18 mm) to test organisms. Fifty-seven isolates formed one cluster, two isolates EBP 2.1 and ZCP 1.9 formed their cluster, while ZCP 17.1 formed a solitary cluster. Among the five test organisms assayed C. albicans formed a single cluster. Correlation between antagonistic activity estimates and bacterial isolates revealed that there were two functional clusters (Figure 6).
16S rRNA analysis
Genomic DNA was extracted from all the 60 bacterial isolates. Partial sequencing for 16S rRNA gene using bacterial specific primers yielded an amplification product of approximately 1500 base pairs. The 60 amplified PCR products were sequenced and only 26 were unambiguous, and their sequences were selected for phylogenetic analyses. The sequences that were >320 base pairs were edited using Chromas pro software and were compared into public databases using BLAST program (http://blast.ncbi.nlm.nih.gov/). Out of 26 isolates submitted to the NCBI database, only 18 were assigned accession numbers MT801052-MT801069 (Table 4). The pairwise alignment was done using MEGA 7 software and the affiliation of the 26 isolates to closest reference strains were determined (Table 4). The phylogenetic relationship of all the partial sequences was determined in MEGA 7 using Maximum-Likelihood analyses. The evolutionary pairwise distances were estimated using the Maximum Composite Likelihood approach (Engeset et al., 2003). The bacterial isolates were identified based on the sequence comparison with the GenBank, NCBI and reference strain. Bacterial isolates were clustered into three different Phyla belonging to Firmicutes, Actinobacteria and Proteobacteria and Bacteriodetes (Figures 7 and 8; Supplementary Table 1). Firmicutes scored 77% closely affiliated with twenty strains, Actinobacteria scored 15% closely affiliated with four strains while Proteobacteria and Bacteriodetes each scored 4% closely affiliated with each strain from both lakes. BLAST analysis of the partial sequences showed there were fourteen isolates (54%) that were closely affiliated with the members of the genus Bacillus with >96 sequence identity from both lakes. Among these were Bacillus group from Lake Olbolosat scoring >97% sequence identity; Bacillus sp recorded three different strains scoring 99.59,100 and 97.43%, B. megaterium scored 99.48%, B. aryabhattai scored 99.89%, B. simplex scored 99.6%, B. subtilis scored 98.84% while B. toyoniensis scored 97.28%. Bacillus group from Lake Oloiden scored >96% sequence identity; two strains of B. pumilus scored 99.69 and 97.26%. Bacillus safensis subsp safensis scored 99.78%, B. altitudinis scored 98.79, Bacillus sp. scored 99.9%, while B. cereus scored 96.45%. Among other Firmicutes were three isolates from the genus Staphylococcus scoring 99.79, 99.78 and 99.59% sequence identities Staphylococcus succinus, S.arlettae and S. xylosus respectively. The other three isolates from the Firmicutes phylum were isolates (ZCP 1.3, ZCP 6.2 and ZAP 10.1) scoring 99.28, 99.54 and 98.84%, sequence identities with known members of the genera Paenibacillus oryzisoli, Bacterium sp and Exiguobacterium sp respectively. Phylum Actinobacteria was affiliated to four different genera Microbacterium oxydans with a score of 99.47%, Streptomyces sp scored 98.56%, Aeromicrobium scored 97.79% while Streptomyces hawaiensis scored 98.73 sequence identities. The Phylum Proteobacteria was closely affiliated with only one strain of Pseudomonas sp. scoring 99.59% sequence identity. There was one strain from the phylum Bacteroidetes closely affiliated to Hymenobacter sp scoring 98.56% (Table 4). There were 10 isolates from Lake Oloiden revealing two clusters. One cluster had strains from Firmicutes with a bootstrap value of 99, while the other one had strains belonging to Actinobacteria with a bootstrap value of 27: one strain formed a node from the latter cluster with a bootstrap value of 9 (Figure 6).
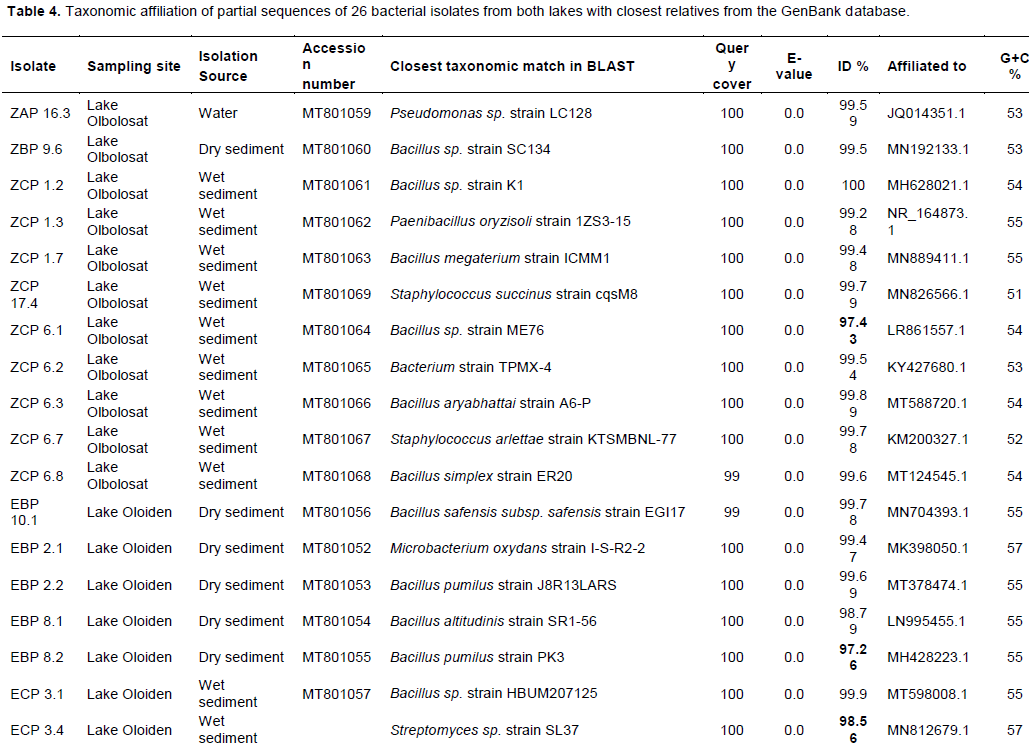
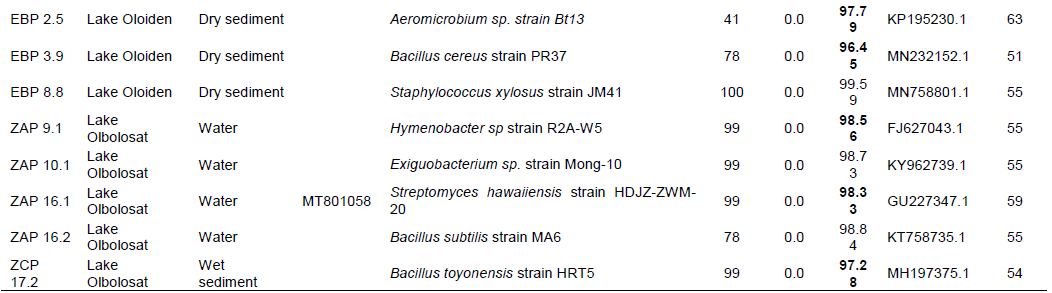

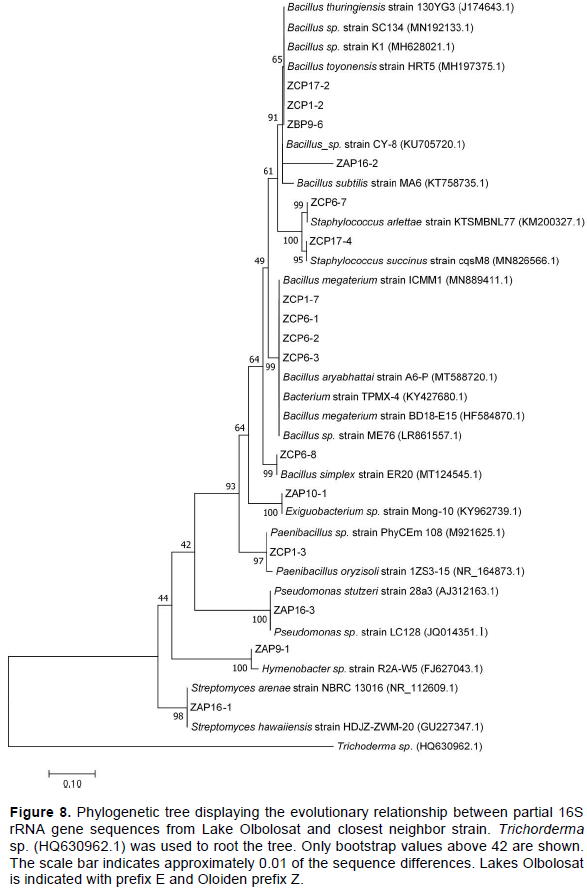
The phylogenetic tree of the 16S rRNA partial sequences of the 16 isolates from Lake Olbolosat revealed two major clusters. One cluster had strains belonging to Firmicutes, proteobacteria and Bacteroidetes with bootstraps values 64, 100 and 100 respectively while the other one had strains belonging to Actinobacteria Phyla with bootstraps values of 98 (Figure 7).
A total of 60 isolates were obtained from lakes Olbolosat and Oloiden. The cfu counts per ml ranged between 0-1.75×105 cfu/ml. The highest cfu counts were obtained from dry sediments within Lake Oloiden while the lowest was from dry sediments from Lake Olbolosat. Growth of isolates in culture medium at different salinity, pH, and temperature ranges indicates that they can tolerate and can adapt to adverse growth conditions in the marine ecosystem. The pH, temperature and salinity are indicators of environmental setting that shapes microbial communities according to O’Brien et al. (2019)and also could affect the activities of extracellular enzymes and breakdown of organic matter (Li et al., 2019). The physiological conditions (pH, temperature, and sodium chloride) are important in the current study if the isolates are to be cultured in the laboratory and be exploited for industrial use (O’Brien et al., 2019).Eight strains from the current study; ZCP 6.1, EBP 8.2, EBP 3.9, ZCP 17.2, ECP 3.4, EBP 2.5, ZAP 9.1 and ZAP 16.1 had sequence similarity of 97.43, 97.26, 96.45, 97.28, 98.56, 97.79, 98.56 and 98.33% sequence similarity respectively representing novel genera of organisms within the lake ecosystem according to Kim et al. (2014)who reported that a bacteria organism could be considered novel if the sequence similarity is <98.65%. The presence of bacteria in the lake ecosystem could be involved in the biodegradation of contaminants such as polycyclic aromatic hydrocarbons through the use of their extracellular enzymes (Yadav et al., 2019). Production of extracellular enzymes by bacterial isolates in this study such as amylases, lipases, proteases, xylanases and cellulases and intracellular enzymes which include starch, catalase, gelatinase and citrase, indicates their biotechnological potential in agriculture, food industries, detergent, medicinal formulations and wastewater management (Yadav et al., 2019). Out of the 60 sequences for bacterial isolates from both lakes, 26 were without ambiguities. The 26 bacterial isolates identified in the current study belonged to the domain bacteria and four different Phyla: Firmicutes, Actinobacteria, Proteobacteria and Bacteroidetes. Firmicutes were predominance within the two lakes. They are known to produce spores that are highly resistant to environmental stress. Formation of spores explains why they may be able to easily outgrow other microorganisms after transfer to microbiological media with their repeated isolation from sediments (Vos et al., 2009). Firmicutes biodegrade complex compounds therefore breaking down macromolecules entering the lake ecosystem such as plants and dead animals providing energy and carbon sources for microbial communities (Vos et al., 2009). There were 13 strains of firmicutes belonging to the family Bacillaceae identified in this study; among these were Bacillus safensis subsp safensis, Bacillus pumilus, B. altitudinis, B. simplex, B. aryabhattai, Bacillus megaterium, B. simplex, B. cereus, B. subtilis and Bacillus toyoniensis. Bacillus pumilus is known to play a good role in the biodegradation of macromolecules in the ecosystem (Mishra et al., 2017). Bacillus altitudinis utilizes various kinds of carbon sources in the lake ecosystem according to Mishra et al. (2017). Bacillus megaterium could be used as an industrial organism since it produces proteins and has been used in bioremediation. Proteins are commonly used in agriculture as plant promoting bacteria and in health sectors (Wafula and Murunga, 2020). Bacillus subtilis, B. cereus, B. pumilus, B. aryabhattai, Bacillus safesis subp safensis and B. simplex have been used as plant promoting bacteria to fix nitrogen, secrete plant hormones or antibiotics, solubilize phosphates, inhibit pathogenic microbes and modify insoluble iron to soluble iron (Chiboub et al., 2018). This is because they are resistant to adverse environmental conditions through the production of spores, they replicate rapidly, and they have a broad-spectrum to biocontrol ability (Chiboub et al., 2018). Plant promoting bacteria are important in enhancing seedling vigor, leaf area, shoot and root growth. Plant promoting hormones like GA3 and IAA are enhanced by the different species of bacteria. The hormone GA3, together with auxin play an important role in the elongation of plant and leaf bud formation (Chiboub et al., 2018). The hormone IAA helps in the emergence and origination of adventitious roots and enhancement of shoot development. Plant promoting hormones also enhances the availability of nutrient uptake to plants helping them against abiotic and biotic stresses (Shafi et al., 2017). The production of antibiotics by bacteria may help them in colonization. Both pathogenic and nonpathogenic organisms compete for space and nutrients with other organisms around them. This is because the soil has a limited amount of nutrients available to sustain them (Shafi et al., 2017). Bacillus cereus occurs naturally and is responsible for most food poisoning (Bartoszewicz and Czyzewska, 2017). Bacillus toyoniensis was isolated from South Africa marine sediment by Ugbenyen et al. (2017)for the production of flocculant used in the biodegrading of pollutants. Staphylococcus spp belonging to Firmicutes occurs ubiquitously in nature and have been isolated from various animals such as birds and mammals (Rossi et al., 2020). Staphylococcus xylosus and S. succinus identified in this study have been used in Italy for the fermentation of traditional sausages (Ratsimba et al., 2017). Staphylococcus xylosus produces biosurfactant an important bioactive compound used in food, cosmetic, petroleum, medicine and pharmaceuticals industries (Ratsimba et al., 2017). Exiguobacterium genus is another Firmicute that was identified in this study and has been isolated earlier from different environmental niches such as sediments, seawater, soils glaciers, hydrothermal vents and industrial effluents (Kasana and Pandey, 2018). Isolates from Exiguobacterium genus can grow under extreme environment with temperature ranging from 12-50°C and under low nutrients conditions (Vishnivetskaya et al., 2009). Different strains from Exiguobacterium genus has been used in industries, in agriculture as a plant promoting bacteria and in biodegradation of pollutants (Kasana and Pandey, 2018). Actinobacteria are known to produce extracellular enzymes and secondary metabolite products. Members of this group are known to have high mol% G+C because of their triple hydrogen bond of the chromosomal DNA content (Hamid et al., 2020). Streptomyces sp, Microbacterium oxydans and Aeromicrobium identified in this study having a high mol% G+C content could make them to adapt to the unfavorable environment, with low mutation rate and tolerant to antagonism factors (Hamid et al., 2020). Streptomyces spp is known to produce 80% of the antibiotic compounds according to Hamid et al. (2020)which are the most important secondary metabolites of the bacterial isolate. M. oxydans and most species in this genus inhabit diverse environments and are associated with the aquatic plants as symbionts according to (Mishra et al., 2017)M. oxydans are also used in a commercial application such as food colorants, dietary supplements, cosmetics and pharmaceuticals purposes (Meddeb-Mouelhi et al., 2016). Bacteroidetes and Proteobacteria are abundant during or following an algal bloom (Meddeb-Mouelhi et al., 2016). Hymenobacter sp. belongs to phylum Bactroidetes was also identified in this study by Royo-Llonch et al. (2017)who reported that Hymenobacter spp, inhabit different environmental niches like marine, fresh water, air, soil, and glacier. Pseudomonas sp a Proteobacteria identified currently is common in the aquatic environment according to (Mishra et al., 2017)and most strains are known to be phosphate solubilizing bacteria and also produce antagonism to other pathogens (Paul and Sinha, 2016).
The study shows that both lakes harbor diverse and novel bacterial species: Firmicutes (Bacillus, Stapylococcus and Exiguobacterium), Proteobacteria (Pseudomonas), Actinobacteria (Streptomyces, Microbacterium, Aeromicrobium) Bacteroidetes (Hymenobacter). The above- mentioned species have the potential for industrial application based on the enzyme, physiological, biochemical, antimicrobial and molecular properties. The study also showed some isolates could be novel strains; ZCP 6.1, EBP 8.2, EBP 3.9, ZCP 17.2, ECP 3.4, EBP 2.5, ZAP 9.1and ZAP 16.1 according to Kim et al. (2014) who reported that a bacteria organism could be considered novel if the sequence similarity is <98.65%. DNA-DNA hybridization could be done to establish the novel strains. An upscale for isolates with the industrial application could also be done as a way forward.
The authors have not declared any conflict of interests.
The authors appreciate the International Foundation for Science (IFS) for funding the work (Grant No.1I1_W_036264). They also appreciate Prof Elijah Ateka for providing Molecular and Biotechnology Laboratory, Dr. Maina Mathaara for providing Microbiology laboratory as well as Dr. Robert Nesta, Dr. Josiah Kuja and Richard Rotich for different analysis.
REFERENCES
|
AU (2015). The African Union Commission Agenda 2063. The Africa We Want. African Union 4:201.
|
|
|
|
Bartoszewicz M, Czyżewska U (2017). Spores and vegetative cells of phenotypically and genetically diverse Bacillus cereus sensu lato are common bacteria in fresh water of Northeastern Poland. Canadian Journal of Microbiology 63(12): 939-950.
Crossref
|
|
|
|
|
Cappuccino JG, Sherman N (2014). Microbiology: A laboratory manual. 6th Edition. Pearson education Inc. San Francisco, California 2:15-224.
|
|
|
|
|
Chiboub M, Jebara SH, Saadani O, Fatnassi IC, Abdelkerim S, Jebara M (2018). Physiological responses and antioxidant enzyme changes in Sulla coronaria inoculated by cadmium resistant bacteria. Journal of Plant Research 131(1):99-110.
Crossref
|
|
|
|
|
Das K, Dutta P (2018). Effects of mulching on soil properties and post-harvest quality of mango Cv. Himsagar grown in New Alluvial Zone of West Bengal International Journal of Agriculture, Environment and Biotechnology New Delhi 11:2.
|
|
|
|
|
Duckworth AW, Grant S, Grant WD, Jones BE, Meijer D (1998). Dietzia natronolimnaios sp. nov., a new member of the genus Dietzia isolated from an East African soda lake. Extremophiles 2(3):359-366.
Crossref
|
|
|
|
|
Engeset RV, Udnæs HC, Guneriussen T, Koren H, Malnes E, Solberg R, Alfnes E (2003). Improving runoff simulations using satellite-observed time-series of snow-covered area. Nordic Hydrology 34(4):281-294.
Crossref
|
|
|
|
|
Hamid ME, Mahgoub A, Babiker AJO, Babiker HAE, Holie MAI, Elhassan MM, Joseph MRP (2020). Isolation and identification of Streptomyces spp. from desert and savanna soils in Sudan. International Journal of Environmental Research and Public Health 17(23):1-10.
Crossref
|
|
|
|
|
Hudzicki J (2012). Kirby-Bauer Disk Diffusion Susceptibility Test Protocol Author Information. American Society for Microbiology 2009:1-13.
|
|
|
|
|
Kasana RC, Pandey CB (2018). Exiguobacterium: an overview of a versatile genus with potential in industry and agriculture. Critical Reviews in Biotechnology 38(1):141-156.
Crossref
|
|
|
|
|
Kim M, Oh HS, Park SC, Chun J (2014). Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. International Journal of Systematic and Evolutionary Microbiology 64(2):346-351.
Crossref
|
|
|
|
|
Kiunjuri M (2017). Republic of Kenya Ministry of Devolution and Planning Reviewed. Government Printer Nairobi 2(1):1-23.
|
|
|
|
|
Krivtsov V, Arthur S, Buckman J, Kraiphet A, Needham T, Gu W, Gogoi P, Thorne C (2020). Characterisation of suspended and sedimented particulate matter in blue-green infrastructure ponds. Blue-Green Systems 2(1):214-236.
Crossref
|
|
|
|
|
Laxma Reddy, PV, Kavitha B, Kumar Reddy PA, Kim KH (2017). TiO2-based photocatalytic disinfection of microbes in aqueous media: A review. Environmental Research 154:296-303.
Crossref
|
|
|
|
|
Li Y, Sun LL, Sun YY, Cha QQ, Li CY, Zhao DL, Song XY, Wang M, McMinn A, Chen XL, Zhang YZ, Qin QL (2019). Extracellular enzyme activity and its implications for organic matter cycling in Northern Chinese Marginal Seas. Frontiers in Microbiology 10:1-13.
Crossref
|
|
|
|
|
Maina CW, Sang JK, Mutua BM, Raude JM (2018). Bathymetric survey of Lake Naivasha and its satellite Lake Oloiden in Kenya; using acoustic profiling system. Lakes and Reservoirs. Research and Management Journal 23(4):324-332.
Crossref
|
|
|
|
|
Meddeb-Mouelhi F, Moisan JK, Bergeron J, Daoust B, Beauregard M (2016). Structural Characterization of a Novel Antioxidant Pigment Produced by a Photochromogenic Microbacterium oxydans Strain. Applied Biochemistry and Biotechnology 180(7):1286-1300.
Crossref
|
|
|
|
|
Mishra RK, Verma DK, Ravindra Yadav MK, Pradhan PK, Swaminathan TR, Sood N (2017). Bacterial diversity and antibiotic resistance in a wetland of Lakhimpur-Kheri, Uttar Pradesh, India. Journal of Environmental Biology 38(1):55-66.
Crossref
|
|
|
|
|
Mohammad BT, Daghistani HI Al, Jaouani A, Abdel-latif S, Kennes C (2017). Isolation and Characterization of Thermophilic Bacteria from Jordanian Hot Springs Bacillus licheniformis and Thermomonas hydrothermalis Isolates as Potential Producers of Thermostable Enzymes. International Journal of Microbiology 2017:1-12.
Crossref
|
|
|
|
|
Moyes RB, Reynolds J, Breakwell DP (2009). Differential staining of bacteria: Gram stain. Current Protocols in Microbiology 15:1-8.
Crossref
|
|
|
|
|
O'Brien FJM, Almaraz M, Foster MA, Hill AF, Huber DP, King EK, Langford H, Lowe MA, Mickan BS, Miller VS, Moore OW, Mathes F, Gleeson D, Leopold M (2019). Soil salinity and pH drive soil bacterial community composition and diversity along a lateritic slope in the Avon River Critical Zone Observatory Western Australia. Frontiers in Microbiology 10:1486.
Crossref
|
|
|
|
|
Paul D, Sinha SN (2016). Isolation and characterization of phosphate solubilizing bacterium Pseudomonas aeruginosa KUPSB12 with antibacterial potential from river. Annals of Agrarian Sciences 15(1):130-136.
Crossref
|
|
|
|
|
Ratsimba A, Leroy S, Chacornac JP, Rakoto D, Arnaud E, Jeannoda V, Talon R (2017). Staphylococcal ecosystem of Kitoza, a traditional Malagasy meat product. International Journal of Food Microbiology 246:20-24.
Crossref
|
|
|
|
|
Rossi CC, Pereira MF, Giambiagi-Demarvalm M (2020). Underrated Staphylococcus species and their role in antimicrobial resistance spreading. Genetics and Molecular Biology 43(1):1-10.
Crossref
|
|
|
|
|
Roux KH (1995). Optimization and troubleshooting in PCR: PCR methods and applications. Cold Spring Laboratory Press 4:185-194.
Crossref
|
|
|
|
|
Royo-Llonch M, Ferrera I, Cornejo-Castillo FM, Sánchez P, Salazar G, Stepanauskas R, González J M, Sieracki ME, Speich S, Stemmann L, Pedrós-Alió C, Acinas SG (2017). Exploring micro-diversity in novel Kordia sp. (Bacteroidetes) with proteorhodopsin from the tropical Indian Ocean via Single Amplified Genomes. Frontiers in Microbiology 8:1-14.
Crossref
|
|
|
|
|
Saitou N, Nei M (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molecular Biology and Evolution 4(4):406-425.
|
|
|
|
|
Shafi J, Tian H, Ji M (2017). Bacillus species as versatile weapons for plant pathogens: a review. Biotechnology and Biotechnological Equipment 31(3):446-459.
Crossref
|
|
|
|
|
Spini G, Spina F, Poli A, Blieux AL, Regnier T, Gramellini C, Varese GC, Puglisi E (2018). Molecular and microbiological insights on the enrichment procedures for the isolation of petroleum degrading bacteria and fungi. Frontiers in Microbiology 9:2543.
Crossref
|
|
|
|
|
Ugbenyen AM, Dafel J, Akapo CO, Mazibuko X, Simonis JJ, Basson AK (2017). Optimisation of the Bioflocculant produced by Pantoea sp, a novel Bacterium Isolated from Marine Sediment 30(3):3066-3073.
|
|
|
|
|
United Nations General Assembly (2015). Global Sustainable Development Report. Global Sustainable Development Report 2015:202.
|
|
|
|
|
Vinet L, Zhedanov A (2010). A "missing" family of classical orthogonal polynomials. Journal of Physics A Mathematical and Theoretical 44(8).
Crossref
|
|
|
|
|
Vishnivetskaya TA, Kathariou S, Tiedje JM (2009). The Exiguobacterium genus: Biodiversity and biogeography. Extremophiles 13(3):541-555.
Crossref
|
|
|
|
|
Vos P, Garrity G, Jones D, Krieg NR, Ludwig W, Rainey FA, Schleifer K-H, Whitman W (2009). Bergeys Manual of Systematic Bacteriology: The Firmicutes. 2nd Edition. William and Wilkins. Baltimore 2(3):19-1314.
|
|
|
|
|
Wafula EN, Murunga SI (2020). Isolation and Identification of Phosphate Solubilizing and Nitrogen-Fixing Bacteria from Lake Ol'Bolossat Sediments, Kenya. Modern Applied Science Journal 14(10):37.
Crossref
|
|
|
|
|
Yadav AN, Yadav N, Kour D, Kumar A, Yadav K, Kumar A, Rastegari AA, GhoshSachan S, Singh B, Chauhan VS, Saxena AK (2019). Bacterial community composition in lakes. In Freshwater Microbiology: Perspectives of Bacterial Dynamics in Lake Ecosystems. Freshwater Microbiology.
Crossref
|
|