ABSTRACT
Twenty six isolates of Colletotrichum gloeosporioides from anthracnose infected mango fruits were isolated from different places of Tamil Nadu, India and these isolates were identified as C. gloeosporioides by Internal transcribed spacer (ITS) and species specific (CgInt) primers. The sensitivity of C. gloeosporioides isolates to benomyl fungicide were evaluated at five different concentrations viz., 0.5, 1, 2, 5, 10 mg/l through poisoned food technique and were expressed very less to highly sensitiveness to the fungicide. All the twenty six isolates were able to grow at the lowest concentration of 0.5 mg/l benomyl amended medium and the average mycelial growth was 70.70 mm at even days after inoculation. At the highest concentration of 10 mg/l benomyl, only two isolates viz., MCG 7 and 16 were able to grow with the mycelial diameter of 12.00 and 18.00 mm, respectively and 100% inhibition was found in the remaining isolates. In addition, benzimidazole sensitive and resistance β - tubulin gene sequences of TUB 1 and TUB2 were amplified from the benomyl sensitive isolates of C. gloeosporioides. The results indicated the differential resistance or sensitivity to benomyl fungicide against C. gloeosporioides and thereby allowed to identify the variability and diversity of the isolates on regional basis.
Key words: Mango, C. gloeosporioides, benomyl sensitive, β - tubulin gene.
Many diseases are affecting mango, of which anthracnose caused by Colletotrichum gloeosporioides (Penz.) Sacc is the most devastating disease and major constraint in production and export of mango. Several fungicides have been used to manage the mango anthracnose as pre- and post-harvest sprays, including benzimidazoles, triazoles, strobilurins, acid amides, dithiocarbamates and heterocyclic compounds (Fei and Wang, 2004). Hot benomyl dips have been reported for effective control of anthracnose in harvested mango fruit (Kim et al., 2007). Benomyl has been used in agriculture for approximately 30 years and numerous cases of
resistance have been reported, including Colletotrichum spp. on various crops (Whiteside, 1980; Picinini, 1994; Hewitt, 1998). Benzimidazoles are specific inhibitors of microtubule assembly that act by binding to their heterodimeric subunit (Davidse, 1986). Several studies have demonstrated that benzimidazole resistance is associated with point mutations in codon 198 or 200 of the β-tubulin gene in various fungi, including Venturia inaequalis, Botrytis cinerea, Monilinia fructicola, C. gloeosporioides and Mycosphaerella fijiensis (Ma et al., 2003; Peres et al., 2004; Chung et al., 2006; Kim et al., 2007). Other point mutations in the β-tubulin gene (Jung et al., 1992; Cooley and Caten, 1993) as well as in the putative leucine zipper protein CaBEN1 (Nakaune and Nakano, 2007) have also been associated with resistance to benzimidazoles. Maymon et al. (2006) reported that sequence analyses of the β-tubulin genes, TUB1 and TUB2, of five sensitive and five resistant representative isolates of C. gloeosporioides from Limonium spp. revealed that the benomyl resistant isolates had an alanine substitute instead of a glutamic acid at position 198 in TUB2. Benzimidazole resistant isolates of C. gloeosporioides from different crops have been identified, but the molecular characteristics of these resistant isolates are still not well defined (Pei, 1981; Tsai et al., 2006). The molecular identification and monitoring of benzimidazole resistant fungi has been recognized as a useful tool in the study of fungicide resistance (Ma et al., 2003; Canas-Gutierrez et al., 2006). Housekeeping genes including the nuclear β-tubulin genes, which show higher DNA sequence variability, are more extensively used to develop diagnostics for fungi, (Fraaije et al., 2001; Mostert et al., 2006; Aroca et al., 2008). Partial β-tubulin sequences from C. gloeosporioides isolates of mango were amplified (Albertini et al., 1999; Yarden and Katan, 1993) as a molecular tool for the identification of benzimidazole-resistant isolates of C. gloeosporioides. Sanders et al. (2000) evaluated 158 isolates of C. gloeosporioides and reported, 17.7% were resistant to benomyl with 8.5% highly resistant due to mutations in the β - tubulin gene using this information, C. gloeosporioides isolates from mango fruits collected in India were evaluated for benomyl sensitivity and mutations in the β - tubulin gene in the present study.
C. gloeosporioides from mango
Twenty six isolates of C. gloeosporioides collected from different districts of Tamil Nadu, India were evaluated in the study (Table 1). Their pathogenicity, variability in virulence was previously reported by Archana et al. (2014).
Sensitivity of C. gloeosporioides to benzimidazole fungicide – benomyl
Each isolate was cultured on Potato Dextrose Agar (PDA) medium in petri plates at 28±2°C for three days, after that mycelial disc (3 mm diameter) were cut from colony margins and a single disc was transferred to the centre of Petri plate containing PDA medium amended with 0.5, 1, 2, 5 and 10 mg/ml of the active ingredient of benomyl fungicide, which was added to PDA medium after autoclaving. Test cultures were incubated in dark at 28±2°C and then mycelial growth was measured 11 days after inoculation (Chung et al., 2010) and control plate was maintained by growing the isolates in the Petri plate containing PDA medium alone.
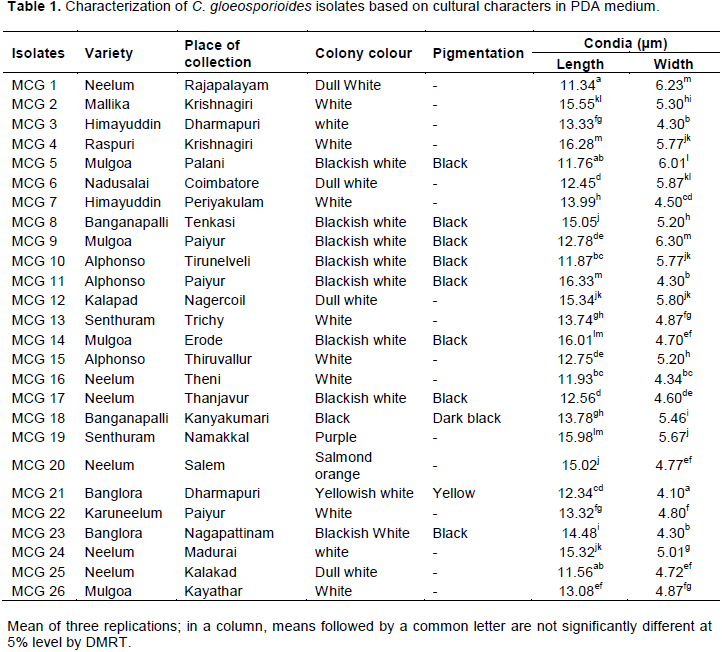
PCR based detection and diagnosis of C. gloeosporioides isolates using internal transcribed spacer (ITS) and CgInt primers
Genomic DNA for C. gloeosporioides isolates were extracted from the mycelial mat by Cetyl trimethyl ammonium bromide (CTAB) method as described by Knapp and Chandlee (1996). The ITS1-5.8S-ITS2 region of ribosomal DNA was amplified with ITS1 (5′-TCCGTAGGTGAACCTGCGG-3′) and ITS4 (5’-TCCTCCGCTTATTGATATGC-3’) oligonucleotide primers. The amplification was performed in 50 μl reaction containing 1.5 units of Taq DNA polymerase (Qiagen, Germany), 1x polymerase chain reaction (PCR) buffer, 200 μM each dNTP, 0.2 μM each primer and 100 ng of template DNA followed by the thermal cycler reaction of 30 cycles (0.5 min at 94°C, 0.5 min at 56°C and 2 min at 72°C) with the final step at 72°C for 7 min (Tosa et al., 2004).
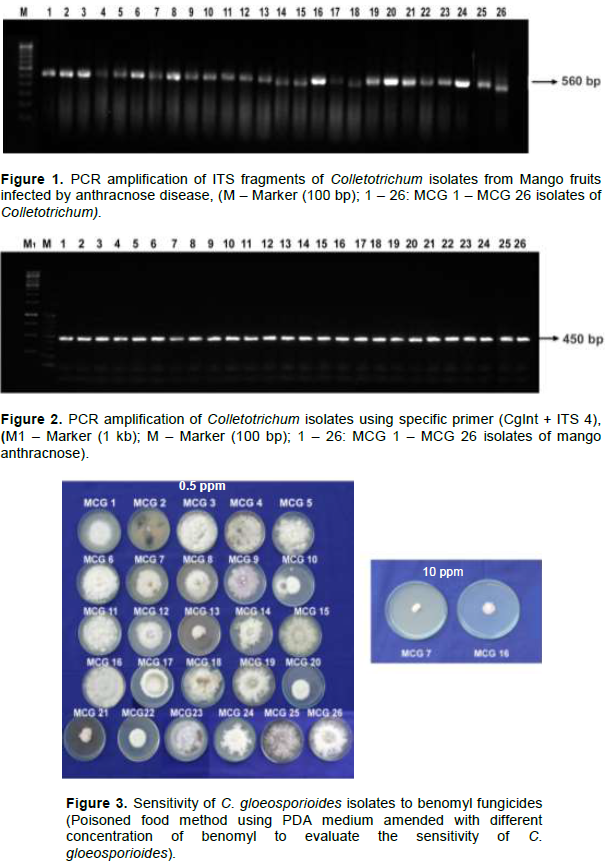
PCR amplification using the oligonucleotide primers ITS 4 (TCCTCCGCTTATTGATATGC) and CgInt (GGCCTCCCGCCTCCGGGCGG) were used for species identification. PCR reaction were as described above with amplification as follows: denaturation at 95°C for 5 min, 30 cycles of 30 s. at 95°C, 30 s at 48°C and 90 s at 72°C and a final extension of 10 min at 72°C (Maymon et al., 2006) in an Eppendorf gradient thermal cycler.
Analysis of benomyl resistance of C. gloeosporioides isolates using partial sequence of β-tubulin gene
PCR ampliï¬cation of β-tubulin gene (TUB2 and TUB2B) fragments were carried out using generic primer GENC (5′-GAGGAATTCCCAGACCGTATGATG- 3′) and TUB2 (5′-GACATCCTTCATAGCG-3′) to identify the benomyl resistant isolates (Koenraadt et al., 1992) and primer GENC and TUB2B (5′-GAC(A/G)TCCTTCAT(A/G)GCGA-3′) to identify benomyl sensitive isolates (Panaccione and Hanau, 1990). TUB1β-tubulin gene was amplified using the primer GENC (5′GAGGAATTCCCAGACCGTATGATG-3′) and TUB 1C (5′-TCAATCTGCTTGGTCGACACCTT-3′) to identify the resistant isolates.
Reactions prepared as above and PCR reactions were performed with 60 s of denaturation at 94°C; followed by 35 cycles consisting of 30 sec at 94°C, 30 sec at 50°C and 60 sec at 72°C; and a final extension period of 10 min at 72°C.
Agarose gel electrophoresis
Amplification products for the above 5 evaluations were separated in 1.2% (w/v) agarose gel in1x TAE buffer (0.4 M Tris, 0.2 M acetic acid, 10mM EDTA; pH 8.4) containing 0.5 µg/ml ethidium bromide. Electrophoresis was carried out at 85 V and the gel was documented using an Alpha Imager (Alpha Innotech Corporation, San Leandro, CA, USA). The sizes of the PCR products were determined by comparing with standard 100 bp or 1 kb molecular marker (Bangalore Genei Pvt. Ltd., Bangalore, India).
Statistical analysis
The data were statistically analyzed using the IRRISTAT version 92 developed by the International Rice Research Institute Biometrics unit, Philippines (Gomez and Gomez, 1984). Data were subjected to analysis of variance (ANOVA) at two significant levels (P< 0.05 and P< 0.01) and means were compared by Duncan’s Multiple Range Test (DMRT).
Molecular detection of C. gloeosporioides isolates using ITS and CgInt primers
PCR amplification using universal primers of ITS 1 and ITS 4 produced the expected 560 bp fragment, confirming that 26 isolates belonged to the genus Colletotrichum (Figure 1). PCR amplification with CgInt and ITS 4 species specific primers produced the expected 450 bp amplicon confirming the isolates were C.
gloeosporioides (Figure 2).
Screening of C. gloeosporioides against benzimidazole fungicide – benomyl
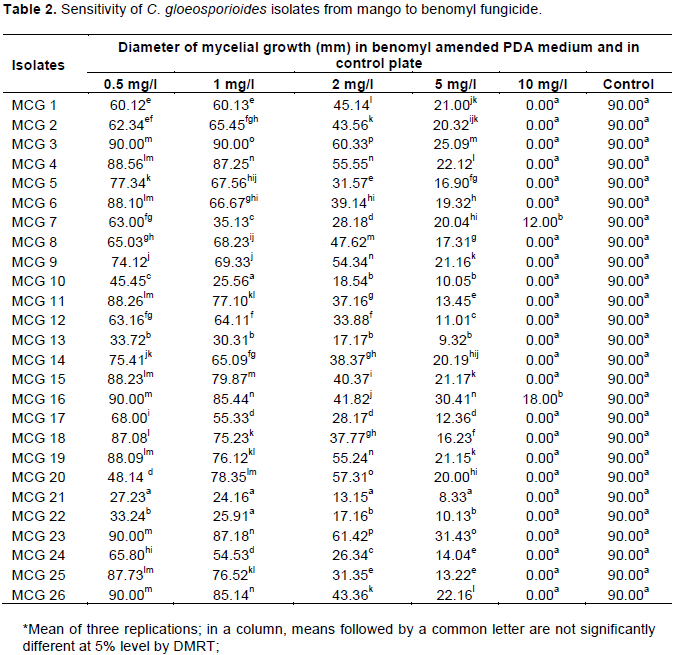
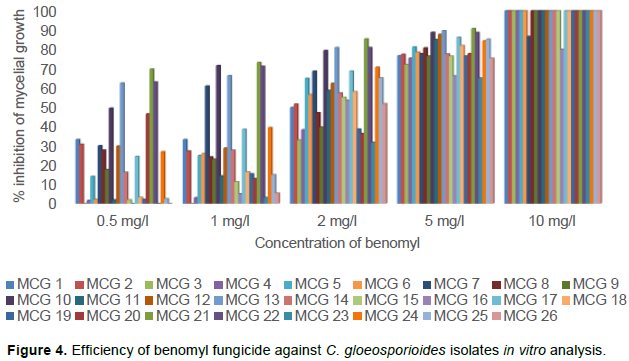
The sensitivity of C. gloeosporioides isolates to the fungicide benomyl was evaluated based on the method of poisoned food technique at five different concentrations viz., 0.5, 1.0, 2.0, 5.0 and 10.0 mg/l (Figure 3). The growth of mycelia on the benomyl amended PDA medium was directly correlated to the sensitiveness of the isolates benomyl. Based on this, twenty six C. gloeosporioides isolates were designated as susceptible, moderately resistant and resistant to the fungicide benomyl. At the lowest concentration of 0.5 mg/l benomyl amended medium, all the twenty six isolates were able to grow and the diameter of the mycelium was measured eleven days after inoculation. The isolate MCG 21 produced the least growth of 27.23 mm followed by MCG 22(33.24 mm) and MCG 13 (33.72 mm) which were significantly different from other isolates. The mycelial growth of 90 mm was observed in the isolates of MCG 3, 16, 23 and 26 at 0.5 mg/l concentration (Table 2). At the concentration of 1 mg/l of benomyl, the percent inhibition over control was higher in the isolate MCG 21 accounting 73.16%, which was on par with the isolates MCG 10 (71.60%) and MCG 22 (71.21%). Least inhibition of 3.06% was observed in the isolate MCG 4. In addition, at the concentration of 2 mg/l the percent inhibition of mycelial growth was ranged from 31.76 to 85.39% and at the concentration of 5 mg/l it ranges from 65.08 to 90.74%. At the highest concentration of 10 mg/l, only two isolates viz., MCG 7 and MCG 16 were able to grow in benomyl poisoned petriplate with the mycelial diameter of 12.00 and 18.00 mm possessing 86.67 and 80.00% inhibition over control, respectively (Figure 4) among 26 isolates. By comparing the mycelial growth of C. gloeosporioides isolates at five different concentrations tested, it was concluded that the isolate MCG 21 was considered sensitive; MCG 7 and MCG 16 were resistant to benomyl fungicide.
PCR amplification of β-tubulin gene from C. gloeosporioides
In all the twenty six isolates of C. gloeosporioides, partial β – tubulin gene sequences of TUB 2 and TUB 1 were amplified
by the appropriate primers from genomic DNA (Figure 5).
The results from PCR amplification, showed that the primer pairs of GENC and TUB2 amplified the resistant allele of TUB 2, (563bp fragment) amplified in ten isolates (MCG 3, MCG 7, MCG 8, MCG 11, MCG 12, MCG 13, MCG 16, MCG 19, MCG 20 and MCG 21) (Figure 5a). The sensitive allele of TUB 2 (563bp fragment) was amplified from nine isolates (MCG 2, MCG 3, MCG 6, MCG 8, MCG 19, MCG 20, MCG 21, MCG 24 and MCG 25) (Figure 5b). In addition, another β-tubulin resistant allele of TUB 1 (506bp fragment) was amplified by the primer pair of GENC and TUB1C from four isolates (MCG 19, MCG 20, MCG 24 and MCG 25) (Figure 5c). Results from this study have confirmed most of the isolates used as benzimidazole-resistant, which were collected from anthracnose infected mango fruits of different places.
Nucleotide sequence accession number
The sequence obtained in this study was assigned GenBank accession number of β-tubulin resistance gene isolates MCG 11-KJ470629.1 and MCG 16-KJ462468.1.
The fungus C. gloeosporioides is known to be highly variable in nature. All twenty six isolates of the present study were identified as C. gloeosporioides based on the symptoms on host, substrate, conidial size and shape, shape of appressoria, growth rate in culture, colour of cultures as reported by von Arx (1970) and Sutton (1980). The traditional method of relying on cultural and morphological grounds for identification of C. gloeosporioidesis variable rarely standardized and tends to mislead, since it is subjective in nature. Molecular genetic studies have provided useful data for clarifying the systematics of the genus Colletotrichum (Martinez et al., 2002). The polymerase chain reaction (Mullis and Faloona, 1987) offers the opportunity to characterize fungal symbionts by amplification of specific sequences and provide very accurate quantitative data required for control and quarantine decisions. Hence, the present study was resorted into exploitation of ITS and CgInt derived primers for identification. The ITS region contains two variable non-coding regions that are nested within the rDNA repeat between the highly conserved small subunit, 5.8S and large subunit rRNA genes (Gardes and Bruns, 1993). The internal transcribed spacer regions (ITS1 and ITS4) within the nuclear ribosomal gene clusters are attractive loci of PCR-based detection assays since they are readily accessible using universal primers (White et al., 1990). In the present study, ITS 1 and ITS 4 primers amplified a fragment of 560 bp corresponding to the region of 18S rDNA sequence in all the twenty six isolates and further confirmed to the group of Colletotrichum. The results confirmed the findings of Kamle et al. (2013) in amplification of ITS region of C. gloeosporioides from mango yielded 560 bp amplicon. Further, identification of ITS regions of nuclear rDNA has been good targets for differentiation and phylogenetic analysis of fungi (Dunne et al., 2002). In the present research, a sensitive PCR-based species-specific designed primer pair of CgInt and ITS 4 was used with the aim to detect C. gloeosporioides of mango anthracnose pathogen from the infected fruit tissue, which amplified the DNA at 450 bp in all isolates. These results were supported by the earlier findings for the taxonomic identification of Colletotrichum species which have been used to distinguish between Colletotrichum acutatum, C. gloeosporioides and Colletotrichum fragariae (Forster and Adaskaveg, 1999; Urena-Padilla et al., 2002). Similarly, Sakinah et al. (2013) reported that, 38 Colletotrichum isolates from banana were identified as C. gloeosporioides (35) and Colletotrichum musae (3) through the amplification of ITS regions and β-tubulin gene using ITS 4 and 5 and Bt2a and Bt2b primers, respectively. Fungicide resistance is a key factor in limiting the efficacy and disease control strategy. The resistance may also be an important aid to our understanding, at a molecular level, of the fungicidal mechanism of action. In the present study, the sensitivity of C. gloeosporioides isolates to benomyl, a benzimidazole fungicide was determined, based on the mycelial growth at five different concentrations viz., 0.5, 1, 2, 5 and 10 mg/l. The twenty six isolates were categorized as three viz., susceptible, moderately resistant and resistant, since significant variation was observed among the isolates in respect to sensitivity. In aqueous solution benomyl is rapidly hydrolysed to methyl benzimidazole – 2 – carbamate and this is probably the active fungi toxicant against pathogen. At the highest concentration of 10 mg/l benomyl, only isolates MCG 7 and MCG 16 were able to grow eleven days after inoculation, which confirmed their sensitiveness of the remaining isolates to benomyl. This might be due to benomyl does not inhibit the spore germination and conidial production but acts primarily by slowing the mycelial growth before or after infection (Peres et al., 2002).
Based on this, isolate MCG 21 was considered as sensitive MCG 5 as moderately resistant and MCG 16 as resistant to benomyl fungicide. This varied reaction obtained in the present study are similar to the findings of Chung et al. (2010), which demonstrated the sensitivity of thirty one C. gloeosporioides isolates from mango and strawberry to benzimidazole fungicides viz., benomyl, carbendazim and thiabendazole. In that, they reported that seventeen isolates of C. gloeosporioides were grouped as sensitive, two isolates were moderately resistant, nine isolates were resistant and three isolates were classified as highly resistant based on the colony diameter on PDA medium amended with benomyl at 10, 100 and 500 mg/ml concentration. It confirms the earlier reports of Sariah (1989), where the growth of sensitive isolates of Colletotrichum capsici were completely inhibited at 2.5 μg/ml benomyl while the resistant isolates grew on agar containing 1000 μg/ml fungicide. Peres et al. (2004) reported that mycelial growth of sensitive isolates was completely inhibited at 1.0 µg/ml of benomyl, whereas resistant isolates grew even at 10 µg/ml of benomyl. Similarly, Joshi et al. (2013) reported that, out of 30 isolates of C. gloeosporioides, isolate Cg 42 from mandarin orange was found to be insensitive to Benomyl with mean IC50 value of 318.85 μg/ml; while Cg 41 and Cg 43 was highly sensitive to Benomyl. In addition Nalumpang et al. (2010) reported the sensitivity of C. gloeosporioides isolates from mango to carbendazim fungicide at various concentrations of 0.1, 1, 10, 100, 500 and 1,000 mg/l. They found that, among 59 isolates of C. gloeosporioides, 49 isolates were highly resistant (HR) phenotypes, 4 isolates were sensitive (S) phenotypes, none showed weakly resistance (WR) and moderately resistance (MR) phenotypes to carbendazim.
Benzimidazole fungicides act by inhibition of tubulin biosynthesis (Davidse, 1973), due to the mutations of β-tubulin gene which has been related to specific amino acid substitutions at several distinct regions within the ß-tubulin molecule (Fujimura et al., 1992). All the twenty six isolates of the current study were used to identify the presence of resistance and susceptible β-tubulin genes. Most often benzimidazole tolerance is due to mutations in the β -tubulin gene which reduce benzimidazole binding (Cooley and Caten, 1993; Reijo et al., 1994). These mutations can be used to rapidly identify tolerant strains with nucleic acid-based methods (Luck and Gillings, 1995).
Further, rapid identiï¬cation of tolerant strains can aid in determining the fungicide resistance management policies. In the present research, partial β - tubulin gene sequences were amplified by the appropriate primers to the DNA fragment for TUB 2 and TUB 1 gene. Isolates MCG 3, MCG 7, MCG 8, MCG 11, MCG 12, MCG 13, MCG 16, MCG 19, MCG 20, MCG 21 had showed amplification product at 563 bp for benomyl resistant gene while the isolates of MCG 2, MCG 3, MCG 6, MCG 19, MCG 21, MCG 24, MCG 25 produced amplification for the primers of β-tubulin sensitive gene. In addition, alternative gene TUB 1 β-tubulin was amplified from four isolates viz., MCG 19, MCG 20, MCG 24 and MCG 25 at 506 bp. Peres et al. (2004) found that eight isolates of C. gloeosporioides were amplified by the portion of β – tubulin gene, with primer TB2R and TB2L.
Resistance to benzimidazole fungicides has been reported in many filamentous fungi, whereby the fungicide is unable to bind to a mutated β-tubulin protein containing an altered amino acid sequence (Davidse and Flach, 1977). Several studies have demonstrated that sequence analysis of the β-tubulin gene of benomyl-resistant fungi revealed that, resistance almost always is due to point mutations in codon 198 or 200 of the β-tubulin gene. Amino acid substitutions at these two codon positions result in different benomyl-resistance phenotypes (Albertini et al., 1999; Yarden and Katan, 1993).
The study reveals that a rapid benomyl tolerance screening could be necessary for examining the potential of different fungicide resistance. Pathogen C. gloeosporioides responsible for anthracnose in mango is composed of both benomyl-resistant and sensitive populations that are genetically distinct, which might be due to a single β-tubulin gene existing within the fungal genome; however, in others, an additional gene may be present that is divergent in sequence.
The authors have not declared any conflict of interests.
This work was supported by the University Grants Commission, Government of India, New Delhi, India, Grant - No. F.14-2 /2010/ (SA-III).
REFERENCES
|
Albertini C, Michel G, Pierre L (1999). Mutations of the β-tubulin gene associated with different phenotypes of benzimidazole resistance in the cereal eyespot fungi Tapesia yallundae and Tapesia acuformis. Pesticide Biochemistry and Physiology 64:17-31.
Crossref
|
|
|
|
Archana S, Prabakar K, Raguchander T (2014). Virulence variation of Colletotrichum gloeosporioides (Penz.) and evaluation of varietal susceptibility against mango anthracnose. Trends in Bioscience 7:415-421.
|
|
|
|
|
Aroca A, Raposo R, Lunello P (2008). A biomarker for the identification of four Phaeoacremonium species using the beta-tubulin gene as the target sequence. Applied Microbiology and Biotechnology 80:1131-1140.
Crossref
|
|
|
|
|
Canas-Gutierrez GP, Patino LF, Rodriguez-Arango E, Arango R (2006). Molecular characterization of benomyl-resistant isolates of Mycosphaerella fjijensis, collected in Colombia. Journal of Phytopathology 154:403 - 405.
Crossref
|
|
|
|
|
Chung WH, Ishii H, Nishimura K, Fukaya M, Yano K, Kajitani T (2006). Fungicide sensitivity and phylogenetic relationship of anthracnose fungi isolated from various fruit crops in Japan. Plant Disease 90:506-512.
Crossref
|
|
|
|
|
Chung WH, Wen-Chuan C, Mun-Tsu P, Hong-Ren Y, Jenn-Wen H (2010). Specific detection of benzimidazole resistance in Colletotrichum gloeosporioides from fruit crops by PCR-RFLP. New Biotechnology 27:17-24.
Crossref
|
|
|
|
|
Cooley RN, Caten CE (1993). Molecular analysis of the Septoria nodorum β-tubulin gene and characterization of a benomyl-resistant mutation. Molecular Genetics and Genomics 237:58-64.
|
|
|
|
|
Davidse LC (1973). Antimitotic activity of methyl benzimidazole-2-yl carbamate (MBC) in Aspergillus nidulans. Pesticide Biochemistry and Physiology 3:317-325.
Crossref
|
|
|
|
|
Davidse LC (1986). Benzimidazole fungicides: mechanisms of action and biological impact. Annual Review of Phytopathology 24:43-65.
Crossref
|
|
|
|
|
Davidse LC, Flach W (1977). Differential binding of benzimidazole2-yl carbamate to fungal tubulin as a mechanism of resistance to this antimitotic agent in mutant strains of Aspergillus nidulans. Journal of Cell Biology 72:174-193.
Crossref
|
|
|
|
|
Dunne CP, Glen M, Tommerup IC, Shearer BL, Hardy GE (2002). Sequence variation in the rDNA ITS of Australian Armillaria species and intra-specific variation in Armillaria. Australasian Plant Pathology 31:241-251.
Crossref
|
|
|
|
|
Fei WC, Wang YM (2004). Plant protection manual. Taiwan Agriculture Chemicals and Toxic Substances Research Institute, Wufeng, Taichung, 835p.
|
|
|
|
|
Forster H, Adaskaveg JE (1999). Identification of sub populations of Colletotrichum acutatum and epidemiology of almond anthracnose in California. Phytopathology 89:1056 -1065.
Crossref
|
|
|
|
|
Fraaije BA, Lovell DJ, Coelho JM, Baldwin S, Hollomon DW (2001). PCR-based assays to assess wheat varietal resistance to blotch (Septoria tritici and Stagonospora nodorum) and rust (Puccinia striiformis and Puccinia recondita) diseases. European Journal of Plant Pathology 107:905-917.
Crossref
|
|
|
|
|
Fujimura M, Kamakura T, Inoue H, Inoue S, Yamaguchi I (1992). Sensitivity of Neurospora crassa to benzimidazoles and N-phenylcarbamates: Effect of amino acid substitutions at position 198 in beta-tubulin. Pesticide Biochemistry and Physiology 44:165-173.
Crossref
|
|
|
|
|
Gardes M, Bruns TD (1993). ITS primers with enhanced specificity for basidiomycetes – application to the identification of mycorrhizae and rusts. Molecular Ecology 2:113-118.
Crossref
|
|
|
|
|
Gomez KA, Gomez AA (1984). Statistical Procedure for Agricultural Research. John Wiley and Sons, New York.
|
|
|
|
|
Hewitt HG (1998). Fungicide resistance. In: Fungicides in crop Protection. Hewitt, H.G. (Ed.) Centre for Agriculture and Biosciences International. Wallingford, UK. pp. 155-181.
|
|
|
|
|
Joshi MS, Sawant DM, Gaikwad AP (2013). Variation in fungi toxicant sensitivity of Colletotrichum gloeosporioides isolates infecting fruit crops. Journal of Agriculture and Food Sciences 3:6-8.
|
|
|
|
|
Jung MK, Wilder IB, Oakley BR (1992). Amino acid alterations in the benA (beta-tubulin) gene of Aspergillus nidulans that confers benomyl resistance. Cell Motility and the Cytoskeleton 22:170-174.
Crossref
|
|
|
|
|
Kamle M, Pradeep K, Vijai KG, Ajay KT, Ashok KM, Brijesh KP (2013). Identification and phylogenetic correlation among Colletotrichum gloeosporioides pathogen of anthracnose for mango. Biocatalysis and Agricultural Biotechnology 2:285-287.
Crossref
|
|
|
|
|
Kim YS, Min JM, Kang BK, Bach NV, Choi WB, Park EW, Kim HJ (2007). Analyses of the less benzimidazole-sensitivity of the isolates of Colletotrichum spp. causing the anthracnose in pepper and strawberry. Plant Pathology Journal 23:187-192.
Crossref
|
|
|
|
|
Knapp JE, Chandlee JM (1996). RNA/DNA mini-prep from a single sample of orchid tissue. Biotechniques 21:54-56.
Crossref
|
|
|
|
|
Koenraadt H, Somerville SC, Jones AL (1992). Characteristics of mutations in the beta-tubulin gene of benomyl - resistant field strains of Venturiainaequalis and other plant pathogenic fungi. Phytopathology 82:1348 - 1354.
Crossref
|
|
|
|
|
Luck JE, Gillings MR (1995). Rapid identiï¬cation of benomyl resistant strains of Botrytis cinerea using the polymerase chain reaction. Mycological Research 99:1483- 1488.
Crossref
|
|
|
|
|
Ma Z, Yoshimura MA, Michailides TJ (2003). Identification and characterization of benzimidazole resistance in Monilinia fructicola from stone fruit orchards in California. Applied and Environmental Microbiology 69:7145 -7152.
Crossref
|
|
|
|
|
Martinez CPV, Barrio E, Suarez-Fernandez MB, Garcia-Lopez MD, Querol A (2002). RAPD analysis of Colletotrichum species isolated from strawberry and the design of specific primers for the identification of C. fragariae. Journal of Phytopathology 150:680-686.
Crossref
|
|
|
|
|
Maymon M, Zveibil A, Pivonia S, Minz D, Freeman S (2006). Identification and characterization of benomyl-resistant and -sensitive populations of Colletotrichum gloeosporioides from statice (Limonium spp.). Phytopathology 96:542-548.
Crossref
|
|
|
|
|
Mostert L, Groenewald JZ, Summerbell RC, Gams W, Crous PW (2006). Taxonomy and pathology of Togninia (Diaporthales) and its Phaeoacremonium anamorphs. Studies in Mycology 54:1- 113.
Crossref
|
|
|
|
|
Mullis KB, Faloona FA (1987). Specific synthesis of DNA in vitro via a polymerase-catalysed chain reaction. Methods in Enzymology 155:335-350.
Crossref
|
|
|
|
|
Nakaune R, Nakano M (2007). Benomyl resistance of Colletotrichum acutatum is caused by enhanced expression of β-tubulin 1 gene regulated by putative leucine zipper protein CaBEN1. Fungal Genetics and Biology 44:1324 -1335.
Crossref
|
|
|
|
|
Nalumpang S, Miyamoto Y, Miyake C, Izumi Y, Akitmitsu K, Kongtragoul P (2010). Point mutations in the beta-tubulin gene conferred carbendazim-resistant phenotypes of Colletotrichum gloeosporioides causing 'Nam Dok Mai' mango anthracnose. Journal of Agricultural Science and Technology 6:365-378.
|
|
|
|
|
Panaccione DG, Hanau RM (1990). Characterization of two divergent beta-tubulin genes from Colletotrichum graminicola. Gene 86:163-170.
Crossref
|
|
|
|
|
Pei CL (1981). Investigation on resistance of phytopathogenic fungi to fungicides in Taiwan. Master Thesis, National Chung Hsing University, 23p.
|
|
|
|
|
Peres NAR, Souza NL, Peever TL, Timmer LW (2004). Benomyl sensitivity of isolates of Colletotrichum acutatum and Colletotrichum gloeosporioides from citrus. Plant Disease 88:125-130.
Crossref
|
|
|
|
|
Peres NAR, Souza NL, Zitko SE, Timmer LW (2002). Activity of benomyl for control of postbloom fruit drop of citrus caused by Colletotrichum acutatum. Plant Disease 86:620-624.
Crossref
|
|
|
|
|
Picinini EC (1994). Fungicides benzimidazoles. Revisão Anual de Patologia de Plantas 2:357-409.
|
|
|
|
|
Reijo RA, Cooper EM, Beagle GJ, Huffaker TC (1994). Systematic mutational analysis of the yeast β-tubulin gene. Molecular Biology of the Cell 5:29-43.
Crossref
|
|
|
|
|
Sakinah MAI, Suzianti IV, Latiffah Z. 2014. Phenotypic and molecular characterization of Colletotrichum species associated with anthracnose of banana (Musa spp.) in Malaysia. Genetics and Molecular Research 13:3627-3637.
Crossref
|
|
|
|
|
Sanders GM, Korsten L, Wehner FC (2000). Survey of fungicide sensitivity in Colletotrichum gloeosporioides from different avocado and mango production areas in South Africa. European Journal of Plant Pathology 106:745-752.
Crossref
|
|
|
|
|
Sariah M (1989). Detection of benomyl resistance in the anthracnose pathogen, Colletotrichum capsici. Journal of Islamic Academy of Science 2:168-171.
|
|
|
|
|
Sutton BC (1980). The Coelomycetes. Commonwealth Mycological Institite. Kew.
|
|
|
|
|
Tosa Y, Hirata K, Tamba H, Nakagawa S, Chuma I, Isobe C, Osue J, Urashima AS, Don LD, Kusaba M, Nakayashiki H, Tanaka A, Tani T, Mori N, Mayama S (2004). Genetic constitution and pathogenicity of Lolium isolates of Magnaporthe oryzae in comparison with host species-specific pathotypes of the blast fungus. Phytopathology 94:454-462.
Crossref
|
|
|
|
|
Tsai JN, Ann PJ, Hu CY, Cheng SF (2006). Evaluation of fungicides for suppression of mycelial growth and conidial germination of Colletotrichum species isolated from mango, pomelo and banana fruit. Plant Pathology Bulletin 15:39-54.
|
|
|
|
|
Urena-Padilla AR, MacKenzie SJ, Bowen BW, Legard DE (2002). Etiology and population genetics of Colletotrichum spp. causing crown and fruit rot of strawberry. Phytopathology 92:1245-1252.
Crossref
|
|
|
|
|
von Arx JA (1970). A revision of the fungi classified as Gloeosporium. A revision of the fungi classified as Gloeosporium.
|
|
|
|
|
White TJ, Bruns T, Lee S, Taylor J (1990). Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand, D.H., Snisky, J.J. and White, T.J. (Eds.) PCR protocols: a guide to methods and applications. pp. 315- 322.
|
|
|
|
|
Whiteside JO (1980). Tolerance of Mycosphaerella citri to benomyl in Florida citrus groves. Plant Disease 64:300 -302.
Crossref
|
|
|
|
|
Yarden O, Katan T (1993). Mutations leading to substitutions at amino acids 198 and 200 of β-tubulin that correlate with benomyl resistance phenotypes of field strains of Botrytis cinerea. Phytopathology 83:1478-1483.
Crossref
|
|