ABSTRACT
Garlic is one of the most cultivated and medicinal horticultural crops in the world. Its production and productivity is decreased due to different constraints. Knowing the genetic distance will give information to tackle the constraints. This study characterized genetic diversity of garlic germplasm by analysis of eleven Simple Sequence Repeats (SSR) loci in 115 garlic accession. Genomic DNA was extracted from fresh leaf tip using modified Diversity Array Technology (DArT) protocol. Touch down polymerase chain reaction (PCR) was also used to get optimal annealing temperature for each Simple Sequence Repeats (SSR) primers. A number of monomorphic and few polymorphic bands were obtained from eleven SSR markers. The gel pictures show monomorphic bands between genotypes. However, some markers: ASM 080, ASA 20, ASA23, and GB-ASM 053 generated polymorphic bands. Dendrogram of genetic distances amongst all tested genotypes showed three distinct major clusters, and six sub-clusters. This study will guide decision making on introduction of germplasm for enhancing genetic diversity or creation of new variations to improve and widen the genetic base of garlic.
Key words: Garlic, genetic variation, polymorphism.
Garlic is one of the main Allium vegetable crops known worldwide with respect to its production and economic value. After potatoes, cassava, and tomatoes, allium vegetables are the fourth most abundant group of commercially produced vegetables (FAO, 2017). Next to onion, it is also cultivated widely (Brewster, 1994). It is one of the very important non-leguminous vegetable crops and used as a flavoring agents in many foods worldwide. Oligosaccharides, steroidal glycosides, essential oil, flavonoids, anthocyanins, lectins, prostaglandins, fructan, pectin, adenosine, and vitamins are the major phytochemical and nutrients found in garlic bulbs (Arzanlou and Bohlooli, 2010).
In Ethiopia, there is a suitable agro-ecological condition for garlic production (Dessie and Mulat, 2019). Due to the genetic constraints, the productivity of garlic is low in many parts of the world as well as in Ethiopia. This can be attributed to lack of improved varieties and abiotic and biotic factors (Velisek et al., 1997). Garlic improvement in Ethiopia was initiated so many years back with the breeding strategy focusing on collection, introduction, adaptation and selection of superior lines. This may be explained by the fact that most of the commercial garlic cultivars are diploid (2n = 16) and vegetatively propagated crops (Ipek et al., 2003). As genetic diversity is the basic input for breeding programmes (Wang et al., 2016), an understanding of the extent of genetic diversity among Ethiopian garlic germplasms is imperative for garlic breeding. Although several studies have been conducted on the agro-morphological characterization of garlic, there is paucity of information on its genetic variation at the molecular level (Sovova and Sova, 2004). Hence, the need to study genetic diversities of garlic at the molecular level.
This work is aimed at determining the extent of genetic variation among Ethiopian garlic accessions. Results from this study will shed light on the pattern of genetic variation across the garlic germplasm; thus, making the identification, grouping, and conservation of garlic lines easy. The information from this work will guide decision making on introduction of germplasm for enhancement of genetic variation and/or creation of new varieties aimed at improving the genetic base of garlic.
Plant materials
Around 115 garlic germplasm/accession were obtained from Ethiopian Institute of Agricultural Research, Deberzeit Agricultural Research Center at intervals of 2003, 2004, and 2006 G.C, Ethiopia. They were planted in different pots under greenhouse conditions.
DNA extraction
Total genomic DNAs were extracted from fresh leaf samples of garlic using a DArt extraction protocol. A 6-ml aliquot of freshly prepared, well-mixed “fresh buffer solution” was preheated to 65ºC and the tubes were placed in a 65ºC incubator. The plant material was ground in a mortar and pestle under liquid nitrogen to a fine powder. The powder was suspended in 6 ml of “fresh buffer solution” kept at 65ºC, and incubated at 65ºC for at least 1 h. The tubes were inverted at every 20 min or incubated with gentle shaking, then cooled down for 5 min, and 6 ml of a chloroform: isoamyl alcohol (24: 1) mixture was added, mixed well for 30 min, and centrifuged for 20 min, 3000 x g, RT. The overlying water phase was transferred to a fresh tube, and the same volume of ice cold isopropanol was added, followed by inversion of the tube ~ 10 times. The visible nucleic acids were sedimented by centrifugation (30 min., 3000 x g, RT). The supernatant was discarded and the pellet was washed with 2 ml of 70% ethanol (EtOH). After discarding the EtOH, the pellet was dried, and dissolved in 200 – 500 µl of 1 x TE (Diversity Array Technology, 2020). Finally, DNA quantification/quality assessments were done using a Nano Drop spectrophotometer, and analyzed by agarose gel electrophoresis.
PCR condition for SSR screening and genotyping
Eleven (11) SSR markers were identified from prior publications (Doyle and Doyle, 2009; Camila et al., 2012). PCR was conducted with 12-μl final volumes, consisting of 1 × buffer, 40 μl genomic DNA, 25 mM MgCl2, 5 μ M primer, 0.2 mM dNTPs, and 0.1 μ L Taq DNA polymerase. The amplification program was as described by Man et al. (2012). The steps involved an initial denaturation step at 94°C for 3 min followed by 35 cycles of 94°C for 30 s. The specific annealing temperature was 55 s, and 60°C for 60 s, with a final extension at 72°C for 10 min. Some primers were done using touch down PCR.
The whole garlic accessions were genotyped by eleven markers with optimized PCR settings for informative markers. Microsatellite alleles were separated using 3% agarose gel electrophoresis. PCR product /DNA ladder (hyper ladder IV) was mixed with loading dye, gel Red mix (in the ratio of 1000:1) and loaded for separation (Table 1).
The gel picture was documented with appropriate resolution. Allele scoring was done with the help of size marker (Hyper ladder IV) loaded together with the experimental samples and/or expected fragment (amp icon) size of the respective markers.
Diversity analysis
Data analysis was done by measuring allele size using PyElph 1.4 software and Darwin6 software used for measuring dissimilarity using modalities dissimilarities indices.
According to Sokal-Michener,
dij = u/m+u
Where, dij = dissimilarities between units i and j
u = number of unmatching units
m = number of matching units.
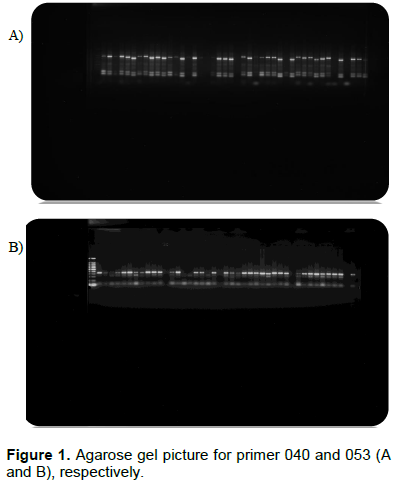
In this study, PCR optimization was performed using all markers using a standard, and touch down PCR programme. A. sativum L. is propagated typically by vegetative means. However, there is a considerable morphological and physiological variation within, and among cultivars (Hirata et al., 2016; Singh et al., 2012). This may be due to the phenotypic plasticity/mutation presence that makes assessment, and systematic classiï¬cation of garlic problematic (Jo et al., 2012). Therefore, analysis of genetic diversity and relatedness between individuals is important for garlic improvement (Kumar et al., 2018). Hoogerheide et al. (2017) also showed that for the purpose of breed improvement, germplasm evaluation from different environments is more reliable for characterization of the genotypic variability between garlic accessions. In this study, there are some markers which show a difference or polymorphism (Figure 1A and B).
Molecular analysis (Figure 2) showed that accessions collected the same year were present together. The present result indicates that molecular diversity is correlated, as observed in three cases, within the same collection year.
In the present study, group of accessions collected from the same location were considered as one population. A number of monomorphic and few polymorphic bands were obtained from 11 SSR markers. The factorial analysis on dissimilarity shows a low Eigen value. Phylogenetic tree of genetic distances among all genotypes showed three distinct major clusters (Figure 2). The major clusters were separated into different sub-clusters. Most of them contain the genotype that was collected in the same year. The dissimilarity value ranged from 0.17 to 0.95.
According to Jo et al. (2012), the diverse eco-geographic conditions of garlic and local selection pressure may be involved in the correlation of molecular diversity within a collection year. Because of clonal propagation nature of the crop, there is a low level of variation as expected.
CONCLUSION AND RECOMMENDATION
Eleven (11) (SSR) primers were used to estimate the genetic diversity and its distribution in 115 garlic genotypes. Most of the gel pictures show the monomorphic band between genotypes. However some markers, ASM 080, ASA 20, ASA23, and GB-ASM 053 generated few polymorphic bands. A dendrogram of genetic distances amongst all tested genotypes, showed three distinct major clusters, and six sub-clusters. This may give good information about genetic diversity among the genotypes but it still needs further data analysis, and interpretation. The genetic diversity of garlic in this work also shows the occurrence of signiï¬cant variability. Therefore, this information indicates that there is a potential of further exploitation in garlic improvement.
The authors have not declared any conflict of interests.
The authors’ appreciate the Ethiopian Institute of Agricultural Research, National Agricultural Biotechnology Research Center (NABRC), for financial support, provision of a greenhouse, and laboratory. The authors are also thankful to the Deberzeit Agricultural Research Center for the garlic germplasm; and National Agricultural Biotechnology Research Center staffs for their unlimited support.
REFERENCES
|
Arzanlou M, Bohlooli S (2010). Inhibition of streptolysin O by allicin an active component of garlic. Journal of Medical Microbiology 59:1044-1049.
Crossref
|
|
|
|
Brewster JL (1994). Onions and other vegetable Alliums. CAB International, Wallingford, UK (Vol. 15).
|
|
|
|
|
Camila PC, Lia EÁ, Hoogerheide SS, Zucchi MI, Monteiro M, Pinheiro JÉB (2012). New Microsatellite Markers for Garlic, Allium Sativum (Alliaceae). American Journal of Botany E17-E19.
Crossref
|
|
|
|
|
Dessie G, Mulat G (2019). Performance of garlic cultivars under rain-fed cultivation practice at South Gondar Zone, Ethiopia. African Journal of Agricultural Research 14(5):272-278.
Crossref
|
|
|
|
|
Diversity Array Technology (2020). Plant DNA Extraction Protocol for DArT.
View
|
|
|
|
|
Doyle JJ, Doyle JL (1990). Isolation of plant DNA fresh tissue. Focus (San Francisco, California) 12:13-15.
|
|
|
|
|
Hirata S, Abdelrahman M, Yamauchi N, Shigyo M (2016). Diversity evaluation based on morphological, physiological and isozyme variation in genetic resources of garlic (Allium sativum L.) collected worldwide. Genes and Genetic System 91:161-173.
Crossref
|
|
|
|
|
Hoogerheide E.S.S, Azevedo Filho J.A, Vencovsky R, Zucchi M.I, Zago B.W, Pinheiro JB (2017). Genetic variability of garlic accessions as revealed by agro-morphological traits evaluated under different environments. Genetics and Molecular Research 16(2):16029612.
Crossref
|
|
|
|
|
Ipek M, Ipek A, Simon P (2003). Comparison of AFLPs, RAPD markers, and isozymes for diversity assessment of garlic and detection of putative duplicates in germplasm collections. Journal of American Society for Horticultural Science 128:246-252.
Crossref
|
|
|
|
|
Jo MH, Ham I, Moe KT, Kwon SW, Lu FH, Park YJ, Kim WS, Won MK, Kim T, Lee EM (2012). Classiï¬cation of genetic variation in garlic (Allium sativum L.) using SSR markers. Australian Journal of Crop Science 6:25-631.
|
|
|
|
|
Kumar M, Sharma VR, Kumar V, Sirohi U, Chaudhary V, Sharma S, Saripalli G, Naresh RK, Yadav HK, Sharma S (2018). Genetic diversity and population structure analysis of Indian garlic (Allium sativum L.) collection using SSR markers. Physiology and Molecular Biology of Plants 25(2):377:386.
Crossref
|
|
|
|
|
Ma KH, Kwag JG, Zhao W, Dixit A, Lee GA, Kim HH, Chung IM (2009). Isolation and characteristics of eight novel polymorphic microsatellite loci from the genome of garlic (Allium sativum L.). Journal of Scientia Horticulturae 122:355-361.
Crossref
|
|
|
|
|
Man HJ, In KH, Kyaw TM, Soon WK, Fu HL, Yong JP, Woon SK, Mi KW, Tae IK, Eun ML (2012). Classification of genetic variation in garlic (Allium sativum L.) using SSR markers. Australian Journal of Crop Science 6(4):625-631.
|
|
|
|
|
Singh RK, Dubey BK, Gupta RP (2012). Studies on variability and genetic divergence in elite lines of garlic (Allim sativum L.). Journal of Spices Aromatic Crops 21:136-144.
|
|
|
|
|
Sovova M, Sova P (2004). Pharmaceutical importance of Allium sativum L. 5. Hypolipemic effects in vitro and in vivo (in Czech). Journal of Ceska Slov Farm 53(3):117-123.
|
|
|
|
|
Velisek J, Kubec R, Davidek J (1997). Chemical composition and classification of culinary and pharmaceutical garlic based products. Z Lebensem Unters Forsch Journal A. 24(2):161-164.
Crossref
|
|
|
|
|
Wang H, Li, X, Liu X, Oiu Y, Song J, Zhang X (2016). Genetic diversity of garlic (Allium sativum L.) germplasm from China by fluorescent based AFLP, SSR and In Del markers. Plant Breeding 135:743-750.
Crossref
|
|