Full Length Research Paper
ABSTRACT
Bacterial wilt caused by Ralstonia solanacearum is one of the most destructive and widespread diseases of tomato in Kenya. The objective of this study was to determine the combining ability effects and gene action conditioning bacterial wilt disease resistance in tomato. Eight parents were crossed in North Carolina II mating design scheme to produce sixteen F1 hybrids. The F1 hybrids and the parental genotypes were evaluated for bacterial wilt in an alpha lattice design. Among the parents, KLF acc III was the best general combiner for area under the disease progress curve (AUDPC) and disease incidence across the two cropping cycles. Red Diamond × KLF acc III, Money Maker× KK acc I, Oxyly× KLF acc III and Money Maker× KK acc II were the best specific combiners for AUDPC. Low narrow sense heritability values of 0.14, 0.16 and 0.20 were obtained for AUDPC, disease incidence and plant survival. Relative weights of additive versus non-additive gene action obtained for AUDPC, disease incidence and plant survival were 0.19, 0.20 and 0.50. General predictability ratios (GPR) values of 0.27, 0.29 and 0.50 were obtained for AUDPC, disease incidence and plant survival. These results indicated the predominance of non-additive gene action in governing the traits.
Key words: Disease resistance, bacterial wilt, combining ability, gene action, tomato.
INTRODUCTION
Tomato (Solanum lycopersicon L.) is one of the most widely cultivated vegetables worldwide. The area under production of this vegetable in Kenya has been on the rise due to the increase in demand (FAOSTAT, 2018; Ochilo et al., 2019). The consumption outstrips the demand and this result from low production that cannot meet the need of the population. Further, there is a gap between the actual and potential yield arising from limiting factors such as lack of suitable varieties coupled with inadequate crop management strategies for control of pests and diseases. Bacterial wilt caused by Ralstonia solanacearum has been identified as a major biotic constraint affecting tomato production in Kenya (Laeshita and Arwiyanto, 2017).
Studies carried out on the inheritance of resistance to bacterial wilt in tomatoes reported the significance of both major and minor genes in regulating the resistance.
Identifying genetic loci responsible for resistance traits, linkage analysis and genome-wide association studies (GWAS) have been widely used (St. Clair, 2010). Quantitative genetic resistance controlled by several genes/Quantitative Trait Loci (QTL), shows complex multigenic inheritance, making breeding efforts challenging (Pilet-Nayel et al., 2017). In disease resistance, haplotype association analysis has been used primarily to characterize diversity at a single target locus in diverse germplasm in order to facilitate the fine mapping of genomic regions containing known resistance loci (Krattinger et al., 2013). A single gene was important for control of bacterial wilt resistance in tomato (Grimault et al., 1995; Thakur et al., 2004). In contrast, the resistance of tomato to bacterial wilt was reported to be under the control of QTLs (Ishihara et al., 2012).
The difference in the results has been attributed to the use of different sources of resistance, variations in environmental conditions and different isolates of R. solanacearum species complex (Da-Silvia et al., 2018). The RSSC strains have been classified into the R. solanacearum species complex, which is composed of four major phylotypes classified according to their geographical origins: I (Asia), II (America), III (Africa and the Indian Ocean), and IV (Indonesia, Australia, and Japan) based on analyses of sequence data derived from the internal transcribed spacer (ITS) region between 16S and 23S (Fegan and Prior, 2005). Recently, the RSSC was taxonomically divided into three species, with phylotypes I and III being classified as R. pseudosolanacearum, phylotype II being classified as R. solanacearum, and phylotype IV being classified as R. syzygii (Prior et al., 2015).
Heritability is a quantitative measure of the genetic variance in phenotypic variation and has predictive value in plant breeding. It indicates the extent to which a particular set of morphogenetic traits can be transmitted through successive generations (Waqar-Ul-Haq et al., 2008). Knowledge of heritability has an effect on the selection procedures used by the plant breeder in determining which selection methods would be most beneficial in improving the traits, predicting gain from selection and determining the relative importance of genetic effects (Laghari et al., 2010). Understanding gene action involved in bacterial wilt resistance in tomato would provide a basis for planning a breeding strategy for developing breeding populations that would lead to identification of superior lines through selection. Alleles with a dominant, additive, or deleterious phenotypic effect have a different effect on heritability when they are homozygous or heterozygous. Understanding how heterozygosity and homozygosity affect gene action and interaction will aid in determining whether hybrids or inbred lines should be used as the end product of breeding programs (Fasoula and Fasoula, 1997). Additive gene action is the mode of gene action in which each of two alleles makes an equal contribution to the generation of qualitative phenotypes. Non-additive gene action is the mode of gene action in which one allele is more strongly expressed than the other (Fasoula and Fasoula, 1997). Non-additive gene action was predominant over additive gene action for the control of resistance to bacterial wilt (Singh et al., 2014). In contrast, additive gene action was important in bacterial wilt resistance (Oliveira et al., 1999). Information on combining ability can help to establish an effective breeding programme. Combining ability analyses is important for facilitating the choice of suitable parents for hybridisation (Suvi et al., 2021). However, combining ability analyses and genetic predictions may depend on the test populations as well as the environment (Suvi et al., 2021). Studies on combining ability have been carried out in other diseases of tomato and other crops. For instance, three tomato lines were identified as potential donors for resistance to tomato yellow leaf curl virus disease in a half-diallel mating design (Pandiarana et al., 2015). Parental lines with negative general combining ability (GCA) values and families with negative specific combing ability (SCA) values were selected for breeding for resistance to rice yellow mottle virus disease (Suvi et al., 2021)
Additive, dominance, and interaction effects of genes, genetic variation in quantitative or complex traits can be partitioned into many components. The additive genetic variance is the most important since it accounts for the majority of the association between relatives and the potential for genetic change via natural or artificial selection (Hill et al., 2008). Additive genetic variance occurs when genes have an additive effect on the quantitative trait. This leads in phenotypic deviation from the mean as a result of the inheritance of a particular allele and its relative effect on the phenotype. It quantifies the degree to which individual phenotype differences may be predicted as a result of allelic substitutions additive effects. Non additive genetic variance is linked with dominant gene acts that encompass the influence of recessive alleles at a particular locus (Singh and Singh, 2018).
The North Carolina II mating design has been widely employed in parental hybridisation for population development and investigating the inheritance of important traits of various crops (Acquaah, 2009; Makanda et al., 2010; Oppong-Sekyere et al., 2019). The design, allows a breeder to estimate the General Combining Ability and Specific Combining Ability (Acquaah, 2009). GCA is defined as a genotype's average performance in a series of hybrid combinations. SCA is defined as those instances in which certain hybrid combinations outperform or underperform their parental inbred lines on an average basis (Sprague and Tatum, 1942). On the basis of SCA, observations of the performance of various cross patterns have been used to infer the gene action at work. The high SCA effects observed in crosses where both parents are good general combiners may be attributed to additive × additive gene action (Dey et al., 2014). The high SCA effects derived from crosses between good and poor general combiner parents may be attributed to the good general combiner parent's additive effects and the poor general combiner parent's epistasis effects, which fit the favourable plant attribute (Verma and Srivastava, 2004). High SCA effects manifested by low crosses may be due to a dominance type of non-allelic gene interaction that results in over dominance, rendering the interaction unfixable (Wassimi et al., 1986). Although studies have revealed the significance of both GCA and SCA in key traits of a number crops including quality traits, disease resistance and yield, limited information exists in the estimation of GCA and SCA from crosses between cultivated and with wild species of tomato (Tyagi et al., 2018). Hence, the study focused on understanding the gene action involved in the control of bacterial wilt and its inheritance. Knowledge of inheritance will be handy in developing a breeding strategy for developing bacterial wilt resistant tomato for both greenhouse and field production.
MATERIALS AND METHODS
Experimental site
The experiment was carried out in the greenhouse at Egerton University, Njoro Campus in the Department of Crops, Horticulture and Soils. The site lies approximately at 35°55'58.0’’E and 0°22’11.0’’S and an altitude of 223 8 m above the sea level. The area is situated in the lower highland agro-ecological zone 3 (LH 3) (Jaetzold et al., 2012).
Genetic material
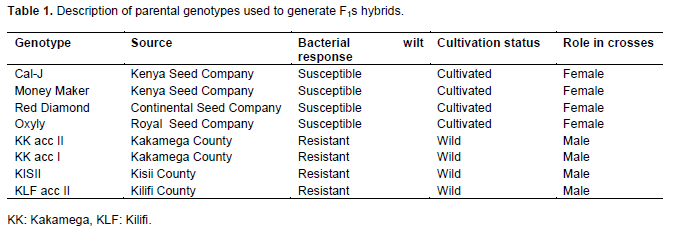
Eight parental genotypes including four commercial susceptible varieties and four wild tomato genotypes with resistance to bacterial wilt were used in the study. Detailed description of these parental materials is provided in Table 1.
Mating parental genotypes
Crossing blocks having eight parents were planted in the greenhouse. Four male parents were crossed to four female parents in North Carolina Design II mating scheme. A total of 16 F1 progenies were obtained. The planting of the parental material was done by staggering to eliminate the possibility of differential flowering time in order to ensure a synchronized flowering period to allow successful crossing. This was achieved by planting the late flowering parents first followed by the early flowering.
Collection, isolation and preservation of R. solanacearum inoculum
Samples of five infected tomato plants showing bacterial wilting symptoms were collected from individual farms in Subukia, Nakuru County in Kenya for isolation of the pathogen. Geographical locations of the farms were recorded using the Global Position System. A quick field ooze test was carried out to distinguish R. solanacearum from vascular wilts that are caused by fungal pathogens. The stems of diseased tomato plants showing typical symptoms of bacterial wilt were cut using sterilized scalpel blades. The cut ends of the stem were placed in test tubes containing sterile distilled water. The presence of the pathogen was confirmed by the proliferation of fine milky white strands when the infected tissue is placed in water. These white strands are as a result of masses of bacteria, which come out of the margins of the cut portions within few minutes (Rohini et al., 2017).
The infected tomato plants collected from the field were washed under running tap water to remove sand and soil. Vascular tissues were extracted with a new sterile scalpel blade into sections of about 10 cm in length from collar region of wilted plants (Ahmed et al., 2013). The tissues were surface sterilized for thirty seconds in 1% sodium hypochlorite solution, 70% ethyl alcohol followed by three repeated washings in sterile distilled water and blot dried. The stem sections weighing one gram were macerated in a test tube containing 10 ml of clean sterile distilled water to create a stock solution. The stock solution was serially diluted by adding 1 ml of bacterial solution to eight test tubes each containing 9 ml of sterile distilled water. Each test tube was vortexed and allowed to settle for at least ten minutes.
Isolation of the bacterium was done following streak plate method as described by Grover et al. (2012) on to 2, 3, 5 Triphenyl Tetrazolium Chloride (Kelman’s TZC agar) medium (glucose 5 g, peptone 10 g, casein hydrolysate 1 g, agar 18 g, distilled water 1000 ml), 5 ml of TZC solution filter sterilized was added to the autoclaved medium to give a final concentration of 0.005%) according to the procedure of Seleim et al. (2014). One loopful of bacterial suspension was obtained from the eight test tubes and streaked on pre sterilized moisture free plates. The plates were incubated upside down in an incubator at 28 ± 2°C for 24-48 h. Single virulent colonies from the medium were characterized by dull white colour fluid with irregular round and light pink centres and these were further streaked on TZC plates to obtain pure culture of the isolates. The pure culture was transferred to 5 mL of sterile double distilled water in screw capped bottles where they were stored for experimental use under refrigeration at -20°C for maintenance of virulence
Experimental procedure
Sixteen F1 alongside eight parents were sowed in a nursery for a period of about 5 weeks before transplanting. The experimental design was an alpha-lattice design of 4 blocks and 6 units within the blocks, in two replicates. The 16F1s with 8 parental genotypes were inoculated with the cultured pathogen 14 days after transplanting. Before inoculation, incisions were made using a sterile scalpel on either side of the main stem to a depth of 5-6 cm each to cause injury to the secondary roots (Mwangi et al., 2008). Thirty millimetres of the standardized bacterial suspension containing 1×109 colony forming units (CFU/ml) per ml inoculation of R. solanacearum was poured over the roots (Singh et al., 2018). Thereafter, the plants were watered at alternative days to maintain a high soil moisture for the development of the disease.
Data collection
All plants in each experimental unit were used for data collection. The disease symptoms were observed daily from 30, 45 and 60 days after inoculation (DAI). The percent disease severity in plants was evaluated using a scale of 0-5 as described by Kempe and Sequeira (1983) (Table 2 and Figure 1).

The disease evaluation data were summarized using the percent
disease severity (PDS) formula as described by Sharma and Saikia (2013) and expressed as the area under the disease progress curve (AUDPC). AUDPC values of 0-150, 151-300, 301-500 and ? 500 were considered to represent very low, low, moderate and high levels of resistance, respectively (Jeger et al., 2001). AUDPC was estimated following Wilcoxson et al. (1975) as:

Data analyses
Data for AUDPC were log transformed while data for disease incidence and plant survival were arcsine square root transformed to obtain a normal frequency distribution. Data were subjected to analysis of variance using the computer software programme GenStat 15th edition (VSN International, Hemel Hempstead, UK). The statistical model for the analysis was;

RESULTS
Analysis of variance and phenotypic performance for AUDPC, disease incidence and plant survival
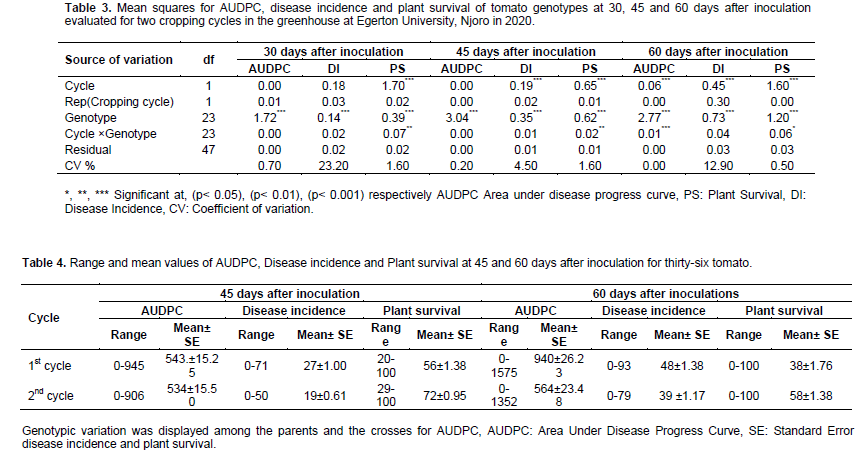
Significant (p < 0.001) variation among the genotypes was recorded across the cropping cycles for AUDPC and plant survival at 30 days and for AUDPC, disease incidence and plant survival at 45 and 60 days after inoculation (DAI) (Table 3). Cropping cycles effects were significant p < 0.001) for plant survival at 30 DAI, disease incidence and plant survival at 45 DAI and AUDPC, disease incidence and plant survival at 60 DAI. Effects due to interaction between genotypes and cropping cycles were significant (p < 0.05) for plant survival at 60 DAI, (p < 0.01) for plant survival at 30 and 45 DAI and (p < 0.001) for AUDPC at 60 DAI.
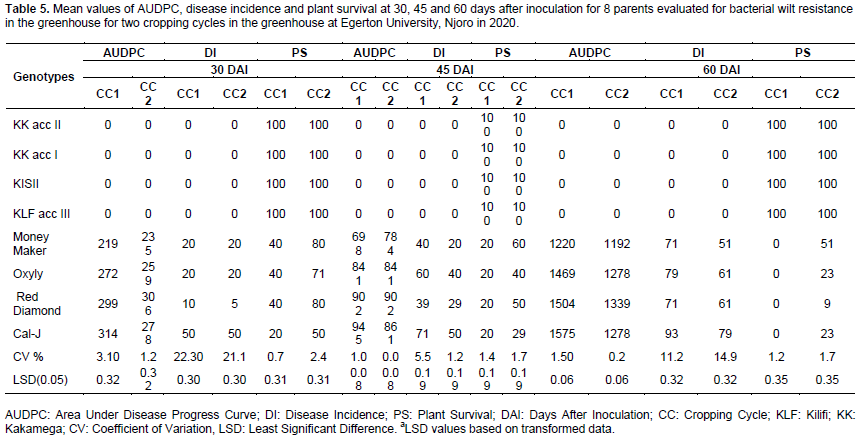
Genotypes expressed variation for AUDPC, disease incidence and plant survival in the two cropping cycles. There was a trend of high disease pressure in the first cropping cycle with mean AUDPC of 543 and 940 at 45 and 60 DAI compared to the second cropping cycle with mean AUDPC of 543 and at 45 and 563 at 60 DAI. In contrast, the plant survival was higher in the second cropping cycle at 45 and 60 DAI with 72 and 58% of the plants surviving compared to the first cropping cycle when only 56 and 38% of the plants survived at 45 and 60 DAI (Table 4).
In general, the crosses recorded lower values for AUDPC and disease incidence and high values of plant survival as compared to the parents. Three crosses Cal-J × KLF acc III, Oxyly × KLF acc III and Red Diamond × KLF acc III and four wild parental genotypes KK acc II, KK acc I, KISII and KLF acc III with AUDPC and disease incidence of 0 values and 100% plant survival were highly resistant compared to commercial varieties which displayed a susceptible reaction to bacterial wilt across cropping cycles (Tables 5 and 6). Apparently all the resistant F1s were progenies of KLF acc III parent.
Combining ability analyses for parents and crosses
Means squares due to parents and crosses were significant (p<0.001) for AUDPC, disease incidence and plant survival. Means squares of Parents× Crosses was significant (p<0.001) for AUDPC and disease incidence. Means squares due to Crosses were significant (p<0.001) for AUDPC, disease incidence and plant survival. Means squares due to Lines× Testers interaction were significant (p<0.001) for AUDPC and disease incidence. Means squares due to Testers was significant (p<0.01) for AUDPC and disease incidence and (p<0.001) for plant survival (Table 7).
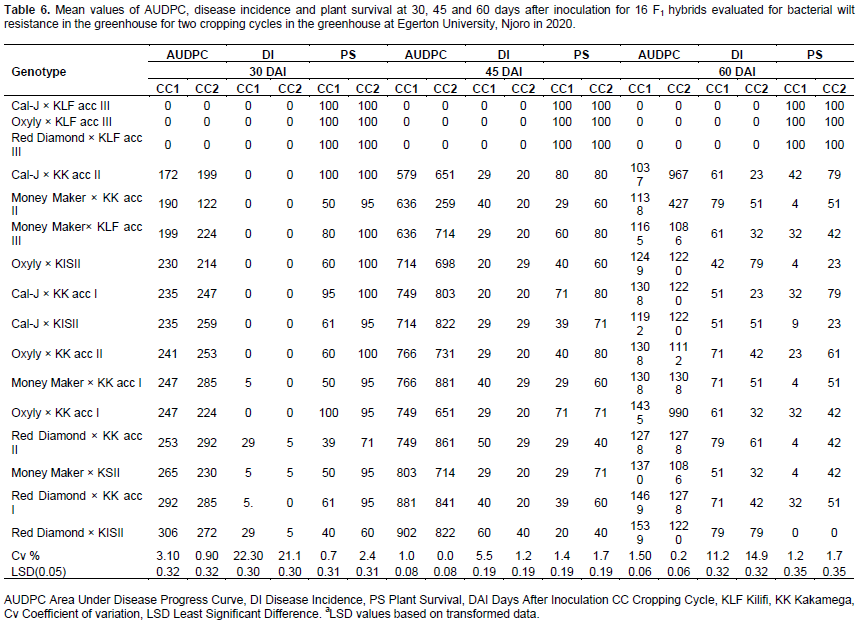
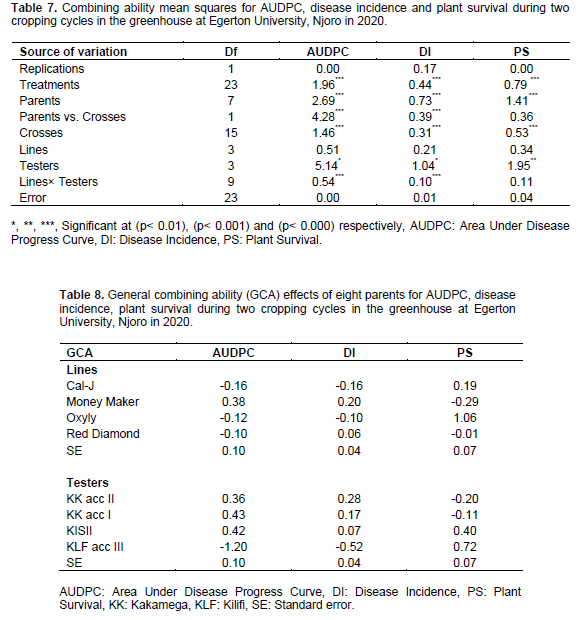
Among the parents, KLF acc III recorded the lowest negative GCA value of -1.20 for AUDPC and -0.52 for disease incidence and high GCA value of 0.72 of plant survival (Table 8). Among the F1s, Red Diamond × KLF acc III, Money Maker× KK acc I, Oxyly× KLF acc III and Money Maker× KK acc II recorded the lowest negative SCA values of -0.41, -0.40 and -0.39. For AUDPC. Red Diamond × KLF acc III recorded the lowest negative SCA value of -0.28 for Disease incidence (Table 9).
Relative weight of additive and non-additive gene action obtained for AUDPC, disease incidence and plant survival were 0.19, 0.20 and 0.50 respectively. Narrow sense heritability values of 0.14, 0.16 and 0.20 were obtained for AUDPC, disease incidence and plant survival. General Predictability Ratios (GPR) values of 0.27, 0.29 and 0.50 were obtained for AUDPC, disease incidence and plant survival. The proportional contribution to the total variation of the testers was higher for all the disease measurements as compared to the lines and the line by testers interaction (Table 10).

DISCUSSION
Bacterial wilt resistance is a major breeding objective for tomato improvement. This is because of the magnitude of yield loss inflicted by the disease which impacts negatively on tomato grown either in the field or under greenhouse conditions. Screening for bacterial wilt resistance has in the past resulted in identification of resistant cultivars (Acharya et al., 2018; Oussou et al., 2020). Despite the existing reports on resistance to bacterial wilt in tomato, local varieties in Kenya are largely susceptible. Introgression of novel sources of resistance from diverse sources including cultivated species and wild relatives is a necessity towards deployment of bacterial wilt resistant tomato cultivars (Kim et al., 2016). Such genetic improvement not only results in reduced yield gap but also helps to reduce production costs and limits the environmental hazards caused by overuse of bactericides.
To determine differential performance among tomato germplasm, AUDPC, disease incidence and plant survival were measured. The results from the analysis of variance revealed the importance of cropping cycle on the performance of tomato against bacterial wilt (Table 3). Significant genotype-by-cropping cycle (GC) interaction for plant survival at 30 and 45 days after inoculation (DAI) and AUDPC and plant survival at 60 DAI suggested that the genotypic performance was not independent of the difference among the cropping cycles. These findings agree with earlier reports (Ganiyu et al., 2017; Guji et al., 2019) and implicate the screening conditions to be key in determining the outcome of disease screening experiment. The variation arising from effects of cropping cycle may result from inconsistent temperature and humidity within the greenhouse. High temperature coupled with high relative humidity accelerate disease development (Velásquez et al., 2018).
Significant main effects due to genotypes for AUDPC, disease incidence and plant survival at 30, 45 and 60 DAI explained the presence of genetic differences among the evaluated genotypes. The trend of higher mean values for AUDPC and disease incidence and lower plant survival at 45 and 60 DAI, observed in the first cropping cycle as opposed to the second cropping cycle suggested higher disease pressure in the second cycle among the genotypes (Table 4). The differential performance may be explained by an increase in temperature during the first cropping cycle. Namisy et al. (2019) found that high temperatures of between 28 to 36°C triggered increased disease pressure.
The observed genetic variation and mean performance of parents and their progenies was based on AUDPC, disease incidence and plant survival which revealed mixed levels of resistance and susceptibility (Tables 5 and 6). Parents with low mean values for AUDPC and disease Incidence and high mean values for plant survival indicated the presence of genes for resistance and the possible potential of transmitting these genes to their progenies (Fellahi et al., 2013). The difference in performance among the parents and the crosses for AUDPC, disease incidence and plant survival indicated the existence of genotypic variation among the parents and the crosses. Suvi et al. (2021) reported genotypic variation for rice yellow mottle virus mottle disease among parents and crosses in rice. Significant mean squares due to testers for the diseases variates suggested the prevalence of additive genetic variance among the male parents in conferring resistance to bacterial wilt (Table 7). These results concur with the earlier findings (Ajjappalavara et al., 2010; Mosa et al., 2017; Kargbo et al., 2019) and therefore indicate that the genetic advance for the disease traits can be realised through hybridisation and selection. Significant mean squares for line × tester interaction for all the traits measured demonstrated the existence of non-additive genetic variance in bacterial wilt resistance. Presence of non-additive genetic variance in the current breeding populations presents the possibility of implementing a hybrid breeding programme that would exploit heterosis in addition to additive gene action to develop new varieties. Tomato hybrids are high yielding and widely cultivated in Kenya and therefore pyramiding resistance genes in inbred lines for deployment of resistant hybrid varieties would greatly improve (Ashkani et al., 2015; Dormatey et al., 2020).
QTL for resistance to tomato late blight was identified in a wild tomato accession (Arafa et al., 2017). QTL linked to bacterial wilt resistance in tomato have been reported by Wang et al. (2018). The QTL identified exhibited a stable and consistent expression. Kumar et al. (2018) identified QTLs linked to bacterial wilt resistance. The QTLs was found to be significantly associated with bacterial wilt resistance. However, bacterial wilt still remains a challenge in tomato production and information on stability of the identified QTLs and their utilization in breeding for resistance is limited. Negative and lower GCA effect for AUDPC and disease incidence recorded by the parent KLF acc III indicated that it was the best general combiner for resistance to bacterial wilt disease (Table 8). Similar findings were reported by Odogwu et al. (2016) bean rust resistance in common bean (Phaseolus vulgaris). The crosses Money Maker× KK acc II, Oxyly× KLF acc III and Red Diamond × KLF acc III recorded negative and lower SCA) effects for AUDPC which showed that these crosses were good specific combiners for resistance to bacterial wilt (Table 9). Bokmeyer et al. (2009) reported that negative SCA effects are desirable for disease resistance.
Heritability is possibly the most important statistic that can be obtained from variance components (Kearsey et al., 1996). Narrow sense heritability measures the proportion of phenotypic variation which arises from additive effects of genes in a given population. Low narrow sense heritability estimates of 0.14, 0.16 and 0.20 obtained for disease traits (Table 10) indicated that dominance gene action was critical in expression of disease resistance for the traits. Low heritability estimates imply that prediction of progeny performance would be difficult because of prevalence of non-heritable variation (Schmidt et al., 2019).
Therefore, a selection procedure that could accumulate positive resistance genes should be adopted. Nsabiyera et al. (2013) reported similar low narrow sense heritability value of 0.16 for bacterial spot. In contrast, Da- Silva Costa et al. (2018) reported narrow sense heritability values of 0.26 and 0.53 for bacterial wilt.
Relative weights of additive and dominance gene action of 0.19, 0.20 and 0.50 respectively for disease traits indicated the superiority of non-additive gene action in their expression (Table 10). Verma and Srivastava (2004) reported the preponderance of non-additive gene action in the expression of traits. General predictability ratio of 0.27, 0.29 and 0.50 for disease traits revealed the predominance of non-additive gene action over additive gene action. This implies that the selection will not be effective and therefore the traits can be improved through use of hybrid vigour. The results are in agreement with Nsabiyera et al. (2013) who reported the predominance of non-additive gene action in the expression of disease traits. In contrast, the inheritance of bacterial wilt has been reported to be controlled by a single dominant gene (Grimault et al., 1995; Thakur et al., 2004). Oliveira et al. (1999) reported additive gene action for resistance to bacterial wilt. Monma et al. (1997) reported the inheritance of bacterial wilt to be partially recessive. Sharma and Sharma (2015) reported the genetic control of bacterial wilt to be oligogenic. In addition, Da- Silva Costa et al. (2018) reported the predominance of additive gene action in the expression of bacterial wilt. Da- Silva Costa et al. (2018) reported the predominance of additive gene action in the expression of bacterial wilt. The proportional contribution of lines, testers and their interaction for the disease traits indicated that testers played an important role in inheritance of disease resistance. The testers contributed more positive alleles for the disease traits (Kargbo et al., 2019). Although both the gene action and both general and specific combining ability effects were evidenced, the predominance of non-additive gene action showed the presence of heterozygosity among the genotypes. From the results, all the four parents were resistant to bacterial wilt. One parent out of four was identified as the best general combiner for bacterial wilt disease. Out of the sixteen crosses, three crosses were resistant to bacterial wilt and had good specific combining ability for bacterial wilt disease resistance. The parent and the three crosses would be useful in tomato breeding programme for the development of a resistant tomato genotypes against bacterial wilt.
CONCLUSION
This study revealed the significance of non-additive gene action in conferring resistance to bacterial wilt. The parental genotype KLF acc III is the best general combiner for bacterial wilt disease. The cross combinations Money Maker× KK acc II, Oxyly× KLF acc III and Red Diamond × KLF acc III had good specific combining ability for resistance to bacterial wilt. From the results, a good breeding strategy would be to concentrate resistance genes in inbred lines with good genetic background through a backcrossing scheme followed by testing for general and specific combing ability for development of hybrids and potential future deployment of genetic resistance in tomato production in Kenya.
CONFLICT OF INTERESTS
The authors have not declared any conflict of interests.
ACKNOWLEDGEMENTS
The authors are grateful for Centre of Excellence in Sustainable Agriculture and Agribusiness Management (CESAAM) project, Egerton University for funding the stud y and appreciate Egerton University for providing institutional support.
REFERENCES
|
Ajjappalavara PS, Dharmatti PR, Salimath PM, Patil RV, Patil MS, Krishnaraj PU (2010). Genetics of bacterial wilt resistance in brinjal. Karnataka Journal of Agricultural Sciences 21(3):424-427. |
|
|
Acharya B, Dutta S, Dutta S, Chattopadhyay A (2018). Breeding tomato for simultaneous improvement of processing quality, fruit yield, and dual disease tolerance. International Journal of Vegetable Science 24(5):407-423. |
|
|
Acquaah G (2009). Principles of plant genetics and breeding John Wiley and Sons. |
|
|
Ahmed NN, Islam MR, Hossain MA, Meah MB, Hossain MM (2013). Determination of races and biovars of Ralstonia solanacearum causing bacterial wilt disease of potato. Journal of Agricultural Science 5(6):86. |
|
|
Arafa RA, Rakha MT, Soliman NEK, Moussa OM, Kamel SM, Shirasawa K (2017). Rapid identification of candidate genes for resistance to tomato late blight disease using next-generation sequencing technologies. PLoS One 12(12):e0189951. |
|
|
Ashkani SMY. Rafii MS, Gous M, Mahbod S, Parisa A, Fatah AT, Mohd SA, Abbas N (2015). Molecular breeding strategy and challenges towards improvement of blast disease resistance in rice crop." Frontiers in Plant Science 6:886. |
|
|
Baker RJ (1978). Issues in diallel analysis. Crop Science 18:533-536. |
|
|
Bokmeyer JM, Bonos SA, Meyer WA (2009). Inheritance characteristics of brown patch resistance in tall fescue. Crop Science 49(6):2302-2308. |
|
|
Da-Silva Costa KD, Dos Santos AMM, Dos Santos PR, Nascimento MR, Silva AMF, Albuquerque GMR, De Carvalho Filho JLS (2018). Inheritance of resistance to Ralstonia pseudosolanacearum in tomato. Euphytica 214(8):1-11. |
|
|
Dey SS, Singh N, Bhatia R, Parkash C, Chandel, C (2014). Genetic combining ability and heterosis for important vitamins and antioxidant pigments in cauliflower (Brassica oleracea var. botrytis L.). Euphytica 195(2):169-181. |
|
|
Dormatey R, Sun C, Ali K, Coulter JA, Bi Z, Bai J (2020). Gene pyramiding for sustainable Crop improvement against biotic and abiotic stresses. Agronomy 10(9):1255. |
|
|
Fasoula DA, Fasoula VA (1997). Gene action and plant breeding. Plant Breeding Reviews 15:315-374. |
|
|
FAOSTAT (2018). Food and Agriculture Organisation of the United Nations. Retrieved December 28, 2018, from FAOSTAT statistics database. |
|
|
Fegan M, Prior P (2005). How complex is the Ralstonia solanacearum species complex. Bacterial wilt disease and the Ralstonia solanacearum species complex 1:449-461. |
|
|
Fellahi ZEA, Hannachi A, Bouzerzour H, Boutekrabt A (2013). Line× tester mating design analysis for grain yield and yield related traits in bread wheat (Triticum aestivum L.). International Journal of Agronomy, 2013. |
|
|
Ganiyu SA, Popoola AR, Enikuomehin OA, Bodunde JG, Adedibu OB, Gurama AU (2017). Assessment of resistance status of some tomato genotypes to bacterial wilt disease and evaluation of SNP marker (LEOH19) for selection of BW resistant gene. Nigerian Journal of Biotechnology 34:54-64. |
|
|
Gashaw G, Alemu T, Tesfaye K (2014). Evaluation of disease incidence and severity and yield loss of finger millet varieties and mycelial growth inhibition of Pyricularia grisea isolates using biological antagonists and fungicides in vitro condition. Journal of Applied Biosciences 73:5883-5901. |
|
|
Grimault V, Prior P, Anais G (1995). A monogenic dominant resistance of tomato to bacterial wilt I Hawaii 7996 is associated with plant colonization by Pseudomonas solanacearum. Journal of Phytopathology 143:349-352. |
|
|
Grover A, Chakrabarti SK, Azmi W, Khurana SMP (2012). Rapid method for isolation of PCR amplifiable genomic DNA of Ralstonia solanacearum infested in potato tubers. Advances in Microbiology 2:441. |
|
|
Guji MJ, Yetayew HT, Kidanu ED (2019). Yield loss of ginger (Zingiber officinale) due to bacterial wilt (Ralstonia solanacearum) in different wilt management systems in Ethiopia. Agriculture and Food Security 8(1):1-11. |
|
|
Hill WG, Goddard ME, Visscher PM (2008). Data and theory point to mainly additive genetic variance for complex traits. PLoS Genetics 4(2):e1000008. |
|
|
Ishihara T, Mitsuhara I, Takahashi H, Nakaho K (2012). Transcriptome analysis of quantitative resistance-specific response upon Ralstonia solanacearum infection in tomato. PLoS One 7(10):me46763. |
|
|
Jaetzold R, Hornetz B, Shisanya, CA, Schmidt H (2012). Farm management handbook of Kenya Vol I-IV (Western Central Eastern Nyanza Southern Rift Valley Northern Rift Valley Coast). Nairobi: Government Printers. |
|
|
Jeger MJ, Viljanen-Rollinson SLH (2001). The use of the area under the disease-progress curve (AUDPC) to assess quantitative disease resistance in crop cultivars. Theoretical and Applied Genetics 102(1):32-40. |
|
|
Jyoti D, Sonia S, Vidyasagar V, Yudhvir S (2015). Inheritance of bacterial wilt resistance and performance of horticultural traits in bell pepper (Capsicum annuum var. grossum). Indian Journal of Agricultural Sciences 85:1498-1503. |
|
|
Kargbo SS, Showemimo F, Akintokun, P, Porbeni J (2019). Combining ability analysis and gene action for yield and yield related traits in rice (Oryza sativa L.) under saline conditions. Journal of Plant Breeding and Genetics 7(2):63-74. |
|
|
Kearsey MJ, Pooni HS, Bulmer M (1996). The Genetical Analysis of Quantitative Traits. Genetical Research 68 (2):183. |
|
|
Kempe J, Sequeira L (1983). Biological control of bacterial wilt of potatoes: attempts to induce resistance by treating tubers with bacteria. Plant disease 67(5):499-503. |
|
|
Kempthorne O (1957). An introduction to genetic statistics. American Psychological Association. https://psycnet.apa.org/record/1958-01083-000 |
|
|
Kim SG, Hur OS, Ro NY, Ko HC, Rhee JH, Sung JS, Baek HJ (2016). Evaluation of resistance to Ralstonia solanacearum in tomato genetic resources at seedling stage. The Plant Pathology Journal 32(1):58. |
|
|
Krattinger SG, Jordan DR, Mace ES, Raghavan C, Luo MC, Keller B, Lagudah ES. (2013). Recent emergence of the wheat Lr34 multi-pathogen resistance: insights from haplotype analysis in wheat, rice, sorghum and Aegilops tauschii. Theoretical and applied genetics 126(3):663-672. |
|
|
Kumar S, Gowda PR, Saikia B, Debbarma J, Velmurugan N, Chikkaputtaiah C (2018). Screening of tomato genotypes against bacterial wilt (Ralstonia solanacearum) and validation of resistance linked DNA markers. Australasian Plant Pathology 47(4):365-374. |
|
|
Laghari KA, Sial MA, Arain MA, Mirbahar AA, Pirzada AJ, Dahot MU, Mangrio SM (2010). Heritability studies of yield and yield associated traits in bread wheat. Pakistan Journal of Botany 42(1):111-115. |
|
|
Laeshita P, Arwiyanto T (2017). Resistance test of several tomato varieties to bacterial wilt diseases caused by Ralstonia solanacearum. Jurnal Perlindungan Tanaman Indonesia 21(1):51-53. |
|
|
Makanda I, Tongoona P, Derera J, Sibiya J, Fato P (2010). Combining ability and cultivar superiority of sorghum germplasm for grain yield across tropical low- and mid-altitude environments. Field Crops Research 116:75-85. |
|
|
Monma S, Sakata Y, Matsunaga H (1997). Inheritance and selection efficiency of bacterial wilt resistance in tomato [Lycopersicon esculentum]. Japan Agricultural Research Quarterly 31(3):195-204. |
|
|
Mosa HE, Abo El-Hares SM, Hassan MAA (2017). Evaluation and Classification of Maize Inbred Lines by Line X Tester Analysis for Grain Yield, Late Wilt and Downy Mildew Resistance. Journal of Plant Production 8(1):97-102. |
|
|
Moussa Z, El-Hersh MS, El-Khateeb AY (2017). Induction of potato resistance against bacterial wilt disease using Saccharomyces cerevisiae. Biotechnology 16(2):57-68. |
|
|
Mwangi JK, Nyende AB, Demo P, Matiru VN (2008). Detection of latent infection by Ralstonia solanacearum in potato (Solanum tuberosum) using stems instead of tubers. African Journal of Biotechnology 7:1644-1649. |
|
|
Namisy A, Chen JR, Prohens J, Metwally E, Elmahrouk M, Rakha M (2019). Screening cultivated eggplant and wild relatives for resistance to bacterial wilt (Ralstonia solanacearum). Agriculture 9(7):157. |
|
|
Nsabiyera V, Ochwo-Ssemakula M, Sseruwagi P, Ojiewo CO, Gibson P (2013). Combining ability for field resistance to disease, fruit yield and yield factors among hot pepper (Capsicum annuum L.) genotypes in Uganda. International Journal of Plant Breeding 7(1):12-21. |
|
|
Ochilo WN, Nyamasyo GN, Kilalo D, Otieno W, Otipa M, Chege F, Lingeera EK (2019). Characteristics and production constraints of smallholder tomato production in Kenya. Scientific African 2:e00014:4. |
|
|
Odogwu BA, Nkalubo S, Rubaihayo P (2016). Genetic analysis of resistance to common bean rust disease in Uganda. RUFORUM Working Document Series (ISSN 1607-9345) 14(1):699-705. |
|
|
Oliveira WF, Giordano LB, Lopes CA (1999). Inheritance of resistance to bacterial wilt in tomato. Fitopatologia 24:49-53. |
|
|
Oppong-Sekyere D, Akromah R, Ozias-Akins P, Laary JK, Gimode D (2019). Heritability studies of drought tolerance in groundnuts using the North Carolina design II fashion and variance component method. Journal of Plant Breeding and Crop Science 11(9):234-253. |
|
|
Oussou GF, Sikirou R, Afoha SA, Dossoumou ME, Boukari SA, Komlan FA, Zocli B (2020). Resistance Assessment of Tomato (Solanum Lycopersicum L.) and Gboma (Solanum Macrocarpon L.) Cultivars Against Bacterial Wilt Caused By Ralstonia Solanacearum in Benin. Pakistan Journal of Phytopathology 32(2):241-249. |
|
|
Pandiarana N, Durwas SV, Seth T, Chatterjee S, Dutta S, Chattopadhyay A (2015). Enhancement of post-harvest fruit quality and leaf curl disease tolerance in tomato through hybrid breeding. Journal of Applied and Natural Science 7(2):606-615. |
|
|
Pilet-Nayel ML, Moury B, Caffier V, Montarry J, Kerlan, MC, Fournet S, Delourme R (2017). Quantitative resistance to plant pathogens in pyramiding strategies for durable crop protection. Frontiers in Plant Science 8:1838. |
|
|
Prior P, Ailloud F, Dalsing BL, Remenant B, Sanchez B, Allen C (2016). Genomic and proteomic evidence supporting the division of the plant pathogen Ralstonia solanacearum into three species. BMC Genomics 17(1):1-11. |
|
|
Team RC (2014). R: A language and environment for statistical computing. Vienna." Austria 2014. |
|
|
Rohini IB, Rangasswamy KT, Achari R (2017). Isolation and characterization of Ralstonia solanacearum causing bacterial wilt of solanaceae crops. International Journal of Current Microbiology and Applied Sciences 6:1173-1190. |
|
|
Schmidt P, Hartung J, Bennewitz J, Piepho HP (2019). Heritability in plant breeding on a genotype-difference basis. Genetics 212(4):991-1008. |
|
|
Seleim MA, Abo-Elyousr KA, Abd-El-Moneem KM, Saead FA (2014). First report of bacterial wilt caused by Ralstonia solanacearum biovar 2 race 1 on tomato in Egypt. The Plant Pathology Journal 30(3):299. |
|
|
Sharma P, Saikia MK (2013). Management of late blight of potato through chemicals. IOSR Journal of Agriculture and Veterinary Science 2:23-36. |
|
|
Sharma KC, Sharma LK (2015). Genetic studies of bacterial wilt resistance in tomato crosses under mid-hill conditions of Himachal Pradesh. Journal of Hill Agriculture 6(1):136-137. |
|
|
Singh S, Singh DR, Kumar K, Birah A (2014). Eco-friendly management modules for bacterial wilt (Ralstonia solanacearum) of tomato for protected cultivation in a tropical island ecosystem. Biological Agriculture and Horticulture 30:219-227. |
|
|
Singh N, Phukan T, Sharma PL, Kabyashree K, Barman A, Kumar R, Ray SK (2018). An innovative root inoculation method to study Ralstonia solanacearum pathogenicity in tomato seedlings. Phytopathology 108(4):436-442. |
|
|
Singh V, Singh K (2018). Additive Genetic Variance. In Vonk J, Shackelford T (eds) Encyclopedia of Animal Cognition and Behavior. Springer, Cham. |
|
|
Sprague GF, Tatum LA (1942). General vs. specific combining ability in single crosses of corn. Journal of the American Society of Agronomy 34(10):923-932. |
|
|
St. Clair DA (2010). Quantitative disease resistance and quantitative resistance loci in breeding. Annual Review of Phytopathology 48:247-268. |
|
|
Suvi WT, Shimelis H, Laing M, Mathew I, Shayanowako AI (2021). Determining the Combining Ability and Gene Action for Rice Yellow Mottle Virus Disease Resistance and Agronomic Traits in Rice (Oryza sativa L.). Agronomy 11(1):12. |
|
|
Thakur AK, Kohli UK, Kumar M (2004). Inheritance of resistance to bacterial wilt in tomato (Lycopersicon esculentum Mill.). Indian Journal of Genetics and Plant Breeding 64(1):79-80. |
|
|
Tyagi V, Dhillon SK, Kaushik P, Kaur G (2018). Characterization for drought tolerance and physiological efficiency in novel cytoplasmic male sterile sources of sunflower (Helianthus annuus L.). Agronomy 8(10):232. |
|
|
Velásquez AC, Castroverde CDM, He SY (2018). Plant-pathogen warfare under changing climate conditions. Current Biology 28(10):R619-R634. |
|
|
Verma OP, Srivastava HK (2004). Genetic component and combining ability analyses in relation to heterosis for yield and associated traits using three diverse rice-growing ecosystems. Field Crops Research 88:91-102. |
|
|
Wang L, Zhou X, Ren X, Huang L, Luo H, Chen, Y, Jiang H (2018). A major and stable QTL for bacterial wilt resistance on chromosome B02 identified using a high-density SNP-based genetic linkage map in cultivated peanut Yuanza 9102 derived population. Frontiers in Genetics 9:652. |
|
|
Waqar-Ul-Haq M, Malik F, Rashid M, Munir M, Akram Z (2008). Evaluation and estimation of heritability and genetic advancement for yield related attributes in wheat lines. Pakistan Journal of Botany 40(4):1699-1702. |
|
|
Wassimi NN, Isleib TG, Hosfield GL (1986). Fixed effect genetic analysis of a diallel cross in dry beans (Phaseolus vulgaris L.). Theoretical and Applied Genetics 72(4):449-454. |
|
|
Wilcoxson RD, Skovmand B, Atif AH (1975). Evaluation of wheat cultivars for ability to retard development of stem rust. Annals of Applied Biology 80:275-28. |
|
Copyright © 2024 Author(s) retain the copyright of this article.
This article is published under the terms of the Creative Commons Attribution License 4.0