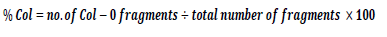ABSTRACT
The disruption of the succinic semialdehyde dehydrogenase (SSADH) function in Arabidopsis leads to aberrant growth phenotype. In human, the SSADH malfunction leads to a condition known as γ-hydroxybutyrate (GHB) aciduria. It is characterized by the accumulation of GHB as well as abnormal growth. GHB aciduria is treated pharmacologically with compounds analogous to gamma-aminubutyrate (GABA). One of these compounds called vagabatrin effectively suppressed the ssadh phenotype in Arabidopsis. Furthermore, the Arabidopsis ssadh phenotype has been suppressed genetically by simultaneous mutation in the POP2 gene. In the present study, the presence of alternative solutions for suppression of the ssadh phenotype was investigated. For that, four ethyl methanesulfonate treated ssadh lines namely 7D, 13J, 17J and 21H were characterized phenotypically and chemotypically. Three- week-old plants of the suppressor lines grew much better than the parent ssadh line and resembled the wild type. The suppressor lines also accumulated reduced amounts of GABA and GHB. PCR based mapping using Sequence Characterized Amplified Region (SCAR) markers located the mutations to chromosome 2 and chromosome 5, suggesting the involvement of at least two mutations in suppression of the ssadh phenotype. Deep whole-genome sequencing of line 17J identified several mutations that induced amino acid change, frame shift and a stop codon. The findings showed that there are more possibilities to suppress the ssadh phenotype; furthermore, it provides an opportunity to identify genes that interact with the GABA shunt.
Key words: Gamma-aminubutyrate (GABA), shunt; succinic semialdehyde dehydrogenase (SSADH) mapping, Arabidopsis, succinic semialdehyde (SSA), γ-hydroxybutyrate (GHB).
Succinic semialdehyde dehydrogenase (SSADH) is a Gamma-aminubutyrate (GABA)-shunt enzyme that catalyzes the oxidation of succinic semialdehyde (SSA) to succinate in the presence of NAD+. Arabidopsis is an annual plant with short life cycle. In plant genetics study, Arabidopsis is often used as a model organism due to its small genome size and short seed to seed cycle. Its genome contains a single copy of SSADH encoding gene (Busch and Fromm, 1999). Knockout of this single gene leads to an accumulation of reactive oxygen species (ROS), a stunted growth (dwarfism), a necrosis on leaves and hypersensitivity to environmental stresses (Fait et al., 2004; Bouche et al., 2003). Furthermore, this abnormal growth phenotype was associated with an accumulation of gamma hydroxybutyric acid (GHB) and hydrogen peroxide in tissues (Fait et al., 2004). In mammals, deficiency in SSADH activity causes a disorder called GHB aciduria, which is manifested by a high level of GHB in the physiological fluids (Jakobs et al., 1981; Hogema et al., 2001).
GHB aciduria in mammals is treated with several drugs. The most commonly used one is g-vinyl-g-aminobutyrate (Vagabatrin), a GABA analogue which binds to GABA transaminase irreversibly and prevents the degradation of GABA (Knerr et al., 2007). A similar rescue of the ssadh phenotype has been reported in Arabidopsis. Arabidopsis ssadh mutant treated with vagabatrin showed a reduced accumulation of ROS, reduced cell death and an improved growth compared to the ssadh mutant (Fait et al., 2004). In 2008, Ludewig et al. (2008) reported a complete suppression of the ssadh phenotype by simultaneous knockout of the POP-2 gene, whose encoded protein functions upstream of the SSADH. This observation suggested that blocking the degradation of GABA is sufficient to suppress the ssadh phenotype.
Functional characterizations of genes are usually done using either forward or reverse genetics approaches. With the reverse genetics approach, one way of mutating genes is by insertion of T-DNA. However, this approach is random and lack specificity. Recent advancements in biotechnological tools allowed targeted edition of genes. CRISPR-Cas technology is one of those technologies that allow targeted modifications of genes (Eid et al., 2016; Schimel et al., 2014). Hyun et al. (2014) used CRISPR-Cas technology to mutagenize FLOWERING LOCUS T and SQUAMOSA PROMOTER BINDING PROTEIN-LIKE genes. Site directed mutagenesis is another method that allows modification of specific amino acids thereby altering the functional properties of proteins. Majeed et al. (2015) and Fontenot et al. (2015) employed site directed mutagenesis to alter single amino acid in UGT76E1 and PhT1 proteins, respectively. The substitution of amino acid threonine (T) by alanine (A) at the position 134 of the UGT76E1 protein abolished the glycosyltransferase activity of the protein (Majeed et al., 2015). Furthermore, the replecement of tyrosine (Y) by Aspartic acid (D) at the 312 position of the PhT1 protein enhanced its Pi transport activity (Fontenot et al., 2015).
Forward genetics approach, on the other hand, involves the induction of random mutation, assessment of phenotypic changes and identification of the mutations responsible for phenotypic change. Several mutagens such as chemicals and rays are used to induce random mutation in the genome of an organism. Ethyl methanesulfonate (EMS) is one of those chemicals which are used as a mutagen in biological studies. EMS induces single nucleotide polymorphisms (SNPs) usually by substitution of bases (Kim et al., 2006). Therefore, EMS mutagenesis has been used widely in forward genetics to identify functions of genes. Previously, Ludewig et al. (2008) isolated two point mutations within the pop2 ORF that suppressed the ssadh phenotype. In the same work, the authors reported the isolation of several EMS suppressor lines. Despite the improved phenotype of the EMS lines, the mode of suppression and genes involved in the process are unknown.
Here, it was hypothesized that mutations in genes other than the POP2 are responsible for suppression of the ssadh phenotype. First, the phenotype and chemotype of four EMS suppressor lines was characterized and named hereafter as 7D, 13J, 17J and 21H. The growth of all four EMS suppressor lines is much better than the parent ssadh plant. The point mutations that suppressed the ssadh phenotype of line 17J were mapped to chromosome 2 and chromosome 5, confirming that mutations in genes other than in POP2 were responsible for suppression of the ssadh phenotype. Deep sequencing of the entire genome of line 17J identified several candidate SNPs that led to amino acid substitution, frame shift and a premature stop codon. Our findings provide a basis for further studies to identify new GABA-shunt interacting genes.
Generation of EMS-suppressor lines
The generation of the EMS suppressor lines was reported previously (Ludewig et al., 2008). Briefly, seeds of ssadh-2 mutant were incubated in water containing 0.23% ethyl methanesulfonate (EMS) for 12 h at room temperature. Then, the seeds were washed several times with water and finally dried. The suppressor lines were isolated by visual observation after sowing the seeds on soil.
Phenotypic analysis
The phenotype lines 7D, 13J, 17J and 21H was compared to wild type and ssadh homozygous plants. For that, seeds of EMS suppressor lines were germinated and grown on soil. After four weeks of growth pictures were taken. Furthermore, the growth of EMS suppressor lines was analyzed on agar plate containing 0.3 mM SSA. For that, seeds of Wt, 7D, 13J, 17J, 21H and pop2 x ssadh mutant (gs) were germinated and grown on ½ MS plate without and with 0.3 mM SSA. Two weeks after germination the fresh shoot weight and root length of plantlets was determined.
Metabolite analysis
The chemotype of lines 7D, 13J, 17J and 21H was compared with the wild type and the ssadh mutant. For that, leaf material (100-200 mg) was collected from four-week-old wild type, gad1/2-ssadh and EMS suppressor lines, and was snap-frozen in liquid nitrogen. To get sufficient material from ssadh mutant, the entire shoots of six-week-old plants excluding flowers were used. Metabolite extraction and measurement was carried out as described previously (Renault et al., 2010; Mekonnen et al., 2016).
Isolation of mapping population
First, line 17J was crossed to Landsberg erecta (Ler) wild type. F1 seeds were sown on soil and the resulting F1 plants were selfed. Next, F2 plants were screened for homozygousity of the T-DNA insertion in the SSADH gene by PCR and also for an improved growth visually. Those lines with homozygous ssadh mutation and improved growth were selected as mapping population. However, this procedure was very laborious, since only few plants tested belonged to the mapping population. Hence, preliminary selection was adopted on agar plates. For that, sterilized F2 seeds were germinated and grown on ½MS plates. A week after germination, only yellowish plantlets were transferred to fresh ½MS plates and after another week of growth the plantlets were transferred to soil. Approximately two weeks after transferring to soil, plants belonging to the mapping population were screened by PCR and visual observation.
Mapping of EMS-induced mutations
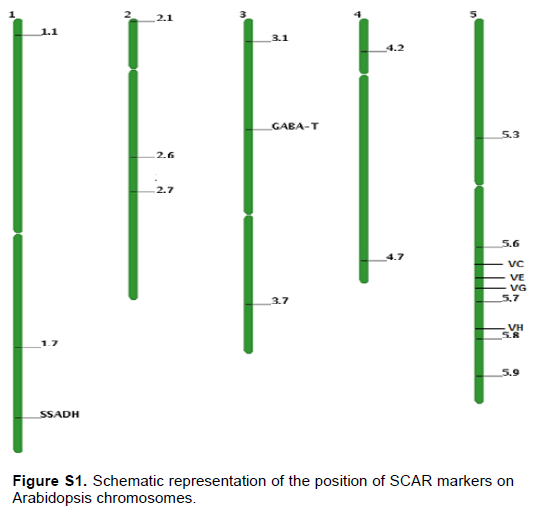
To determine the position of SNPs that suppressed the ssadh phenotype, genomic DNA was extracted from the mapping populations as described previously (Sholz et al., 2015). Markers discriminating between the Columbia (Col-0) and Landsberg erecta (Ler) fragments were designed on all five chromosomes (Figure S1 and Table S1). PCR was performed on genomic DNA extracts of the mapping population. For genomic DNA extraction the procedure described by Scholz et al. (2016) was used. The abundance of Col-0 and Ler fragments was counted as follows. A single band per lane corresponding to Col-0 or Ler is counted as two and double bands per lane counted one for each genotype. The number of bands was summed up and the percent of Col fragment was calculated as shown by the following formula:
Genomic DNA extraction for whole genome sequencing
High quality genomic DNA was extracted from three-week-old 17J line germinated and grown on soil. For that ~100 mg of leaf material was collected from line 17J and immediately frozen in liquid nitrogen. The extraction of the genomic DNA was carried out according to the DNeasy Plant Mini Kit protocol (Cat. No. 69104). The concentration of the DNA was determined using a Nano Drop.
The EMS suppressor lines grow better than the ssadh mutant
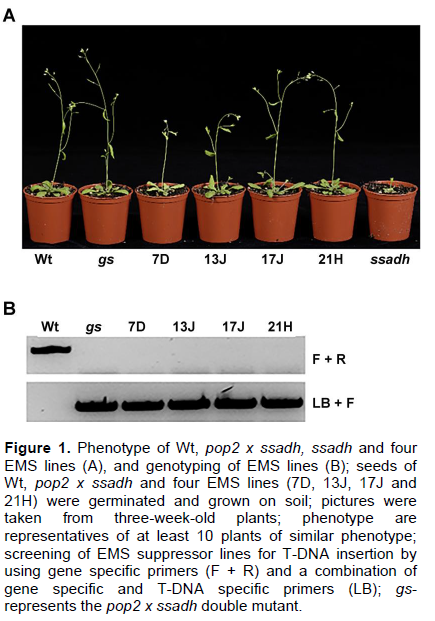
The growth of Arabidopsis ssadh mutants is severely inhibited (Ludewig et al., 2008; Fait et al., 2004). However, the simultaneous knockout of the POP2 gene completely suppressed the ssadh phenotype (Ludwig et al., 2008). In the present study, the ssadh phenotype was suppressed by random mutations induced by EMS. The growth of EMS suppressor lines (7D, 13J, 17J and 21H) was much better than the ssadh mutant (Figure 1A). Three-week-old plants of line 17J and 21H resemble the wild type and pop2 x ssadh mutant, confirming the suppression of the ssadh phenotype (Figure 1A). To ensure that the T-DNA insertion in the SSADH gene was stable, the EMS suppressor lines were genotyped using gene- and T-DNA-specific primer combinations. Indeed, all lines were homozygous for T-DNA insertion; since, the PCR using gene specific forward and reverse primers did not yield a product (Figure 1B).
Furthermore, if EMS suppressor lines would be sensitive to SSA treatments differently compared to the controls was tested. Similar to soil grown plants, there was no difference in shoot growth between the EMS suppressor lines and the controls (Wt and gs) on ½ MS plates (Figure 2A). However, a shorter root was measured in all EMS lines when compared to the wild type on ½ MS plates (Figure 2B). This might be due to the accumulation of GABA after the disruption of the SSADH function. The accumulation of GABA in SSADH deficient Arabidopsis plants has previously been reported (Ludwig et al., 2008; Fait et al., 2004). A high GABA level is associated with reduced root growth. Ramesh et al. (2015) showed a reduced growth of wheat roots after 10 mM exogenous GABA application. Treatment of 0.3 mM SSA reduced the growth of all lines including the wild type (Figure 2C), an observation in line with the previous report (Mekonnen and Ludwig, 2016). The suppression of the ssadh phenotype by EMS induced mutation could be in two ways; first by preventing the production of the toxic intermediates; second, by promoting the degradation of the toxic intermediate(s). The phenotype of EMS lines following SSA treatment probably suggests the first mode of suppression of the ssadh phenotype. The phenotypic difference among EMS lines might indicate the presence of alternative ways to rescue the ssadh phenotype.
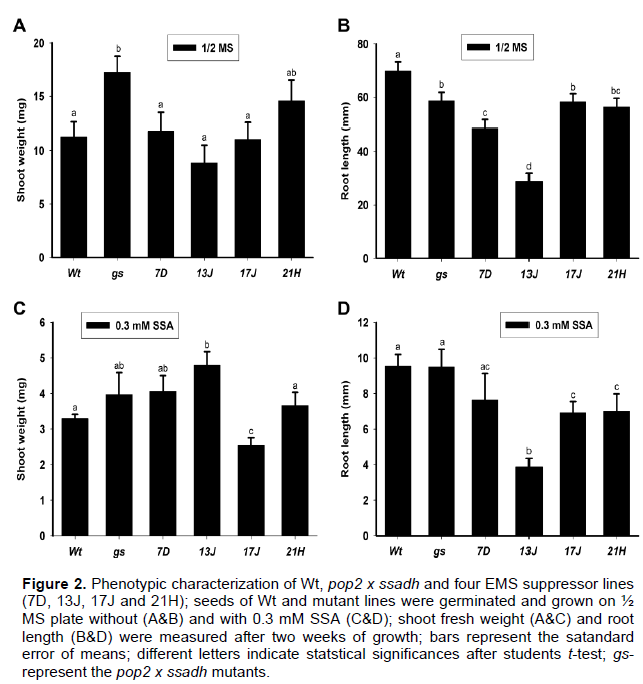
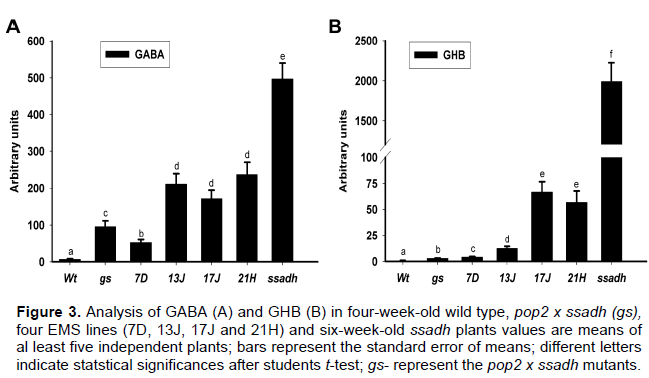
EMS induced mutations reduced the GABA and GHB levels
Some of the features of the ssadh chemotype include the accumulation of high GABA and GHB in tissues (Ludwig et al., 2008). In the present report, the abundance of GHB and its correlation with the suppression an ssadh phenotype was investigated. Interestingly, the GHB content was reduced by more than 30-fold in shoots of EMS suppressor lines compared to the ssadh mutant (Figure 3B). Similarly, the GABA content in EMS suppressor lines was reduced by more than 50% compared to the ssadh mutant (Figure 3A).The reduction in levels of GABA and GHB suggests that prevention of toxic intermediates production was probably the mode of ssadh phenotype suppression. Intriguingly, lines 7D and 13J exhibited a relatively reduced shoot growth phenotype compared to 17J and 21H (Figure 1A) despite accumulating several folds less GHB in the tissue, suggesting that the severity of the ssadh phenotype is not correlated to the GHB level. Akaboshi et al. (2003) reported the absence of correlation between GHB levels and clinical features in mammals. Our recent report also showed that GHB induced damage in Arabidopsis is modest and zero in yeast at high concentrations (Mekonnen and Ludwig et al., 2016). This observation adds further evidence to a previous report that GHB is not the main cause for ssadh phenotype.
SNPs in at least two genes suppressed the ssadh phenotype
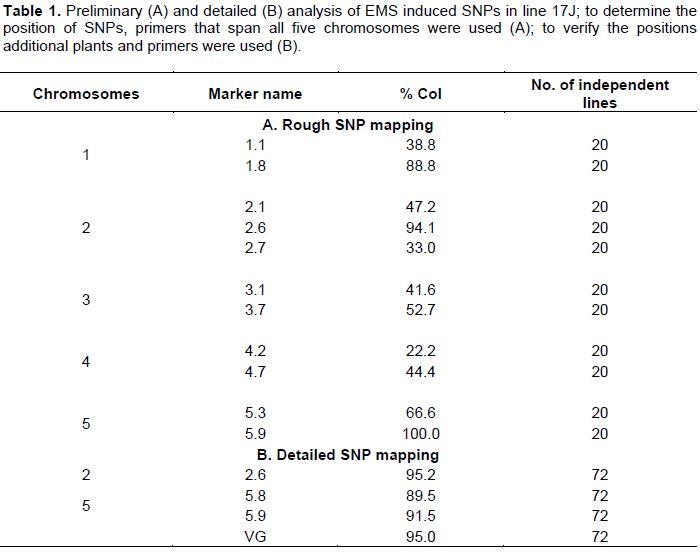
The PCR based mapping system using Sequence Characterized Amplified Region (SCAR) markers has been used widely to locate SNP’s. The presence of natural polymorphisms between two Arabidopsis ecotypes (Col and Ler) lays a good platform for designing SCAR markers. For example, Konieczny and Ausubel (1993) identified an average of one nucleotide change in 261 base pairs between Col and Ler ecotypes that comprised recognition cites for CAPS marker. Based on the nucleotide polymorphisms between these two ecotypes, several SCAR markers were designed spanning the shorter and longer arms of all five chromosomes (Figure 4 and Table S1). Using a combination of PCR genotyping and visual analysis, more than 90 lines belonging to the mapping populations was isolated for line 17J. Interestingly, PCR analysis using markers 1.8, 2.6 and 5.9 identified three regions on chromosome1, 2 and 5, respectively, which is rich in Col-0 fragments (Table 1A). The lower arm of chromosome 1 is a position where the ssadh T-DNA insertion was located. Therefore, it was not surprising to see dominance of Col-0 fragments. To confirm the position of the SNPs, additional PCRs was performed by using more lines from the mapping population and found that the mutations were, indeed, in chromosome 2 and 5 (Table 1B). Those findings show that the ssadh phenotype was suppressed by at least two mutations in two different chromosomes. Despite the absence of a segregation analysis data, the proportion of plants belonging to the mapping population in the F2 generation was close to 1/16.
Deep sequencing of line 17J genome uncovered several candidate mutations
The introduction and accessibility of full genome sequencing facilities greatly eased the identification of SNP’s. The whole genome sequencing of line 17J identified hundreds of point mutations compared to the reference allele. To determine the regions of interest on chromosome 2 and 5, the following calculations were made. Markers 2.6 and VG on chromosome 2 and 5, respectively, yielded a 95% Col-0 fragment and 5% Ler fragment (Table 1B). Centimorgan (cM) is a unit of measuring the linkage between genes on the same chromosome. The 5% Ler fragments in chromosomes 2 and 5 (Table 1B) indicate that five crossing over has occurred between the markers and EMS induced SNP’s. From the above definition, it can be deduced that the markers and EMS induced SNPs are 5 cM apart. Although the conversion of 1 cM to base pair depends on the position on the chromosome, the average number was calculated to be 208950 and 203800 base pairs in chromosome 2 is 5, respectively (http://www.arabidopsis.org/ servlets/mapper). The region of interest was obtained by multiplying the estimated base pairs with 5. To ensure the selection of the right SNPs, a 5% cut off value was set for the ratio of reference calls to alternative calls, that is, any ratio value higher than 0.05 was discarded.
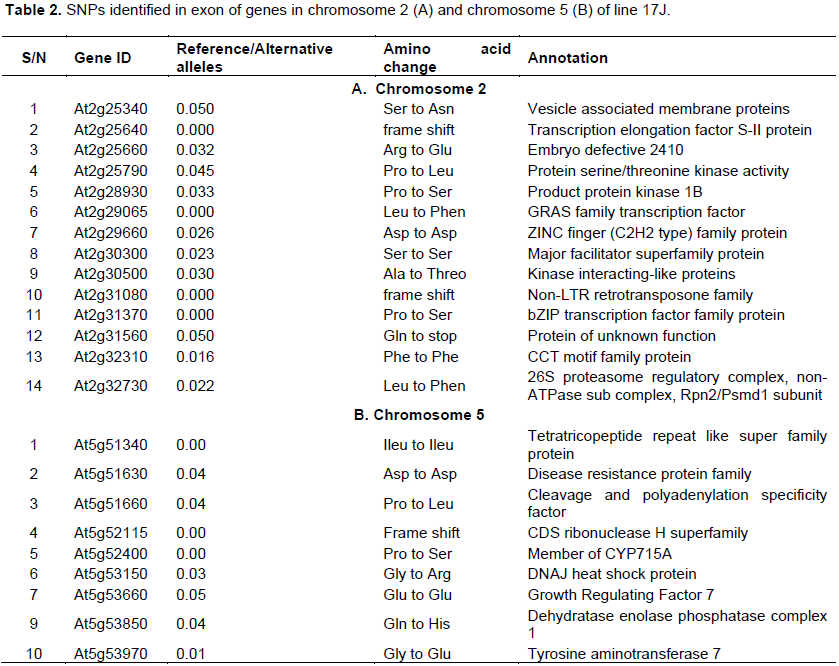
Sequence analysis revealed hundreds of SNPs within the desired region of both chromosomes. However, only 47 and 22 SNPs were true in chromosome 2 and 5, respectively, since more than 95% of the calls favored against the reference allele. The SNPs were located within the exon, intron, 3’ UTR or 5’ UTR of the gene. Among all SNPs, 14 and 10 point mutations in chromosome 2 and 5, respectively, were located within the exon of genes (Table 2). The point mutations induced amino acid changes, premature stop codon and a frame shift (Table 2). Furthermore, there were some notable changes in terms of the amino acid polarity and charge in both chromosomes. For example, the substitution of Gly by Asp, Arg by Glu, Gly by Arg, and Gly by Glu were the notable ones in genes of both chromosomes (Table 2). Such substitution could change the function of the protein. Single amino acid substitutions have previously been shown to alter the functional property of proteins. Lee et al. (2015) reported the enhanced peroxidase and chaperone activity of 2-Cys Peroxiredoxin (2-Cys Prx A) protein following the substation of Cys150 for Ser150. Similarly, the replacement of Thr134 by Ala134 of Arabidopsis UGT76E1 protein abolished its glycosyltransferase activity (Majeed et al., 2015). Such functional alteration of proteins will likely have consequences on the phenotype (Ng and Henikoff,
2006). The suppression of the ssadh phenotype was, indeed, the manifestation of the amino acid change. To our knowledge no report has been published indicating interaction between these candidate genes and the GABA shunt.
The only known way of suppressing of the ssadh phenotype in Arabidopsis is through mutations in POP2 gene. The finding here provided potential candidate genes that have never been reported to interact with the GABA shunt before. The candidate genes can be used to study their role in suppression of ssadh phenotype as well as their mode of interaction with the GABA shunt. Considering the diverse roles of the GABA shunt in plant growth and development, there is a promise of identifying new enzymatic pathways. Furthermore, in humans the disorder resulting from ssadh malfunction is treated only with chemical drugs (GABA-analogues) that competetively inhibit the function of GABA-T (POP2). The identification of additional pathways that suppress the ssadh phenotype is helpful; since, the information could be used as an input for the discovery of drugs in GHB aciduria treatment of mammals. Taken together, the result of the study lays a good basis to identify new genes and possibly pathways that interact with the GABA shunt.
The authors have not declared any conflict of interests.
REFERENCES
|
Akaboshi S, Hogema BM, Novelletto A, Malaspina P, Salomons GS, Maropoulos GD, Jakobs C, Grompe M, Gibson KM (2003). Mutational spectrum of the succinate semialdehyde dehydrogenase (ALDH5A1) gene and functional analysis of 27 novel disease-causing mutations in patients with SSADH deficiency. Hum. Mutat. 22:442-450.
Crossref
|
|
|
|
Bouche N, Fait A, Bouchez D, Moller SG, Fromm H (2003). Mitochondrial succinic- semialdehyde dehydrogenase of the caminobutyrate shunt is required to restrict levels of reactive oxygen intermediates in plants. Proc. Natl. Acad. Sci. 100:6843-6848.
Crossref
|
|
|
|
Busch KB, Fromm H (1999). Plant succinic semialdehyde dehydrogenase. cloning, purification, localization in mitochondria, and regulation by adeninenucleotides. Plant Physiol. 121(2):589-598.
Crossref
|
|
|
|
Eid A, Ali Z, Mahfouz MM (2016). High efficiency of targeted mutagenesis in Arabidopsis via meiotic promoter-driven expression of Cas9 endonuclease. Plant Cell Rep. 35(7):1555-1558.
Crossref
|
|
|
|
Fait A, Yellin A, Fromm H (2004). GABA shunt deficiencies and accumulation of reactive oxygen intermediates: insight from Arabidopsis mutants. FEBS Lett. 579:415-420.
Crossref
|
|
|
|
Fontenot EB, DiTusa SF, Kato N, Olivier DM, Dale R, Lin WY, Chiou TJ, Macnaughtan MA, Smith AP (2015). Increased phosphate transport of Arabidopsis thaliana Pht1;1 by site-directed mutagenesis of tyrosine 312 may be attributed to the disruption of homomeric interactions. Plant Cell Environ. 38(10):2012-2022.
Crossref
|
|
|
|
Hogema BM, Gupta M, Senephansiri H, Burlingame TG, Taylor M, Jakobs C, Schutgens RB, Froestl W, Snead OC, Diaz-Arrastia R, Bottiglieri T (2001). Pharmacologic rescue of lethal seizures in mice deficient in succinate semialdehyde dehydrogenase. Nat. Genet. 29:212-216.
Crossref
|
|
|
|
Hyun Y, Kim J, Cho SW, Choi Y, Kim JS, Coupland G (2014). Site-directed mutagenesis in Arabidopsis thaliana using dividing tissue-targeted RGEN of the CRISPR/Cas system to generate heritable null alleles. Planta 241(1):271-284.
Crossref
|
|
|
|
Jakobs C, Bojasch M, Mönch E, Rating D, Siemes H, Hanefeld F (1981). Urinary excretion of gamma-hydroxybutyric acid in a patient with neurological abnormalities. The probability of the new inborn error of metabolism. Clin. Chim. Acta 111:169-178.
Crossref
|
|
|
|
Kim YS, Schumaker KS, Zhu JK (2006). EMS Mutagenesis of Arabidopsis. Methods in Molecular Biology 323: Humana Press Inc., Totowa, NJ.
|
|
|
|
Knerr I, Pearl PL, Bottiglieri T, Snead OC, Jakobs C, Gibson KM (2007). Therapeutic concepts in succinate semialdehyde dehydrogenase (SSADH; ALDH5a1) deficiency (gamma-hydroxybutyric aciduria). Hypotheses evolved from 25 years of patient evaluation, studies in Aldh5a1-/- mice and characterization of gamma-hydroxybutyric acid pharmacology. J. Inherit. Metab. Dis. 30(3):279-294.
Crossref
|
|
|
|
Konieczny A, Ausubel FM (1993). A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J. 4(2):403-410.
Crossref
|
|
|
|
Lee EM, Lee SS, Tripathi BN, Jung HS, Cao GP, Lee Y, Singh S, Hong SH, Lee KW, Lee SY, Cho JY, Chung BY (2015). Site-directed mutagenesis substituting cysteine for serine in 2-Cys peroxiredoxin (2-Cys Prx A) of Arabidopsis thaliana effectively improves its peroxidase and chaperone functions. Ann. Bot. 116(4):713-725.
Crossref
|
|
|
|
Ludewig F, Hueser A, Fromm H, Beauclair L, Bouche N (2008). Mutants of GABA transaminase (POP2) suppress the severe phenotype of succinic semialdehyde dehydrogenase (ssadh) mutants in Arabidopsis. PLoS One 3(10):e3383.
Crossref
|
|
|
|
Majeed HN, Zia MA, Yang M, Sheikh MA, Bhatti IA (2015). Cloning and site directed mutagenesis of UGT76E1 leads to changed substrate activity in Arabidopsis thaliana. Int. J. Agric. Biol. 17(6):1125-1132.
Crossref
|
|
|
|
Mekonnen DW, Fluegge UI, Ludewig F (2016). gamma-aminobutyric acid depletion affects stomata closure and drought tolerance of Arabidopsis thaliana. Plant Sci. 245:25-34.
Crossref
|
|
|
|
Mekonnen DW, Ludwig F (2016). Phenotypic and chemotypic studies using Arabidopsis and yeast reveal that GHB converts to SSA and induce toxicity. Plant Mol. Biol. 91(4-5):429-440.
Crossref
|
|
|
|
Ng PC, Henikoff S (2006). Predicting the effects of amino acid substitutions on protein function. Annu. Rev. Genomics Hum. Genet. 7:61-80.
Crossref
|
|
|
|
Ramesh SA, Tyerman SD, Xu B, Bose J, Kaur S, Conn V, Domingos P, Ullah S, Wege S, Shabala S, Feijó JA (2015). GABA signaling modulates plant growth by directly regulating the activity of plant-specific anion transporters. Nat. Commun. 6:7879.
Crossref
|
|
|
|
Renault H, Roussel V, El Amrani A, Arzel M, Renault D, Bouchereau A, Deleu C (2010). The Arabidopsis pop2-1 mutant reveals the involvement of GABA transaminase in salt stress tolerance. BMC Plant Biol. 10:1-16.
Crossref
|
|
|
|
Schimel S, Fauser F, Puchta H (2014). The CRISPR/Cas system can be used as nuclease for in planta gene targeting and as paired nickases for directed mutagenesis in Arabidopsis resulting in heritable progeny. Plant J. 80(6):1139-1150.
Crossref
|
|
|
|
Scholz S, Reichelt M, Mekonnen DW, Ludewig F, Mithoefer A (2015). Insect herbivory elicited GABA accumulation in plants is a wound-induced, direct, systemic and jasmonate-independent defense response. Front. Plant Sci. 6:1128.
Crossref
|