Full Length Research Paper
ABSTRACT
Groundnut chlorotic rosette disease (GCRD) transmitted by the aphid, Aphis craccivora, is an important virus disease of groundnut in Africa. Breeding for host resistance remains the best strategy to minimize losses due to this disease. Nine cultivated groundnut genotypes with differential reaction to GCRD were crossed in an incomplete diallel mating design to determine the combining ability of GCRD resistance. The parents and 36 F2 populations were inoculated with veruliferous A. craccivora at the seedling stage and evaluated for disease reaction at two locations in Nigeria in 2012. Disease incidence (based on visual symptoms) was recorded three times at fortnightly interval using area under disease progress curve. General combining ability (GCA) and specific combining ability (SCA) effects for GCRD resistance were highly significant (P < 0.01), indicating that both additive and non-additive gene effects governed the inheritance of GCRD resistance. The Baker ratio was low (0.3) for GCRD indicating that non-additive gene effects was more important than additive gene effects in controlling GCRD resistance in these crosses. As a result, progeny performance could not be adequately predicted from GCA effects alone. Therefore, effective selection of superior genotypes would be achieved at advanced generations when maximum homozygosity is attained.
Key words: Groundnut chlorotic rosette disease, area under disease progress curve, combining ability, additive and non-additive gene effects.
INTRODUCTION
Groundnut (Arachis hypogaea L.), is cultivated annually on about 24.63 million hectares worldwide with annual production of 41.27 million tons in shell with a productivity of about 1.85 t ha–1 (FAO, 2012). It is highly adapted to tropical and subtropical climates of the world and cultivated in nearly 100 countries. It is a key crop for small scale farmers especially in Africa and Asia where the crop serve as a valuable source of dietary protein, oil, and fodder for livestock. It contains 48-50% oil and 26-28% protein, and is a rich source of dietary fibre, minerals, and vitamins (Janila et al., 2013). In addition, groundnut has the ability to fix atmospheric nitrogen to the soil to help in the maintenance of soil fertility. In West Africa, Nigeria is the largest producer of groundnuts with a production of 3.07 million tons on about2.4 million hectares (FAO, 2012). Despite the economic, social and cultural importance of groundnuts, its productivity is severely constrained by several biotic and abiotic factors. Among them groundnut chloroticrosette disease (GCRD) causes severe crop losses The most serious yield losses were reported during the year 1975 when an epidemic in northern Nigeria destroyed approximately 0.7 million hectares of groundnut, with an estimated loss of US$250 million (Yayock et al., 1976).The GCRD is characterised by small, chlorotic, twisted and distorted leaflets with shortened internodes and thickened stems. Affected plants especially those infected at a young stage are severely stunted (Bock et al., 1990). The disease also affects both quality of the haulm and the pod.
Three agents Groundnut rosette assistor virus (GRAV), Groundnut rosette virus (GRV) and a Satellite-RNA (Sat-RNA) (Reddy et al., 1985; Murant et al., 1988; Taliansky et al., 2000) contribute to the etiology. The intimate interaction between GRAV, GRV, and sat-RNA is crucial to the development of the disease.
Host resistance is the most cost effective and environmentally friendly method to minimize losses due to GCRD. Several resistant varieties have been developed in West Africa Resistance to GCRD is not immunity and succumbs to high inoculum pressure and adverse environmental conditions (Bock et al., 1990). Breeding for resistance to groundnut rosette disease demands a good knowledge of the breeding methodologies as well as a good understanding of the disease and its causal organisms. Identification of sources of resistance and its efficient utilization require an understanding of the genetic control of resistance and knowledge of the amount of genetic variability available for selection. Determining the suitable parents to use for the development of resistant genotype is particularly important.Early genetic studies on groundnut rosette disease showed that resistance was effective against GRV and its sat-RNA and was governed by two independent recessive genes (de Berchoux, 1960). He also stated that resistant lines were not immune and that individual plants could become infected when subjected to inoculation by massive number of aphids. This resistance was reported to operate equally against both chlorotic rosette (de Berchoux, 1960) and green rosette (Harkness, 1977). He attributed the low recovery of resistant plants from Virginia x Spanish crosses to heavy inoculum pressure at an early stage of growth and suggested occurrence resistance breakdown from generation to generation. Bock and Nigam (1988) studied the inheritance of resistance to chlorotic rosette (GRV and its sat-RNA) in crosses involving botanical varieties of groundnut from Malawi and confirmed the findings of de Berchoux (1960) of two recessive genes governing the resistance in all the backgrounds. In resistant plants, the presence of GRAV was detected. Gene conferring resistance to GRV and its sat-RNA did not confer resistance to GRAV (Bock and Nigam, 1988; Bock et al., 1990). Similar findings on the inheritance of resistance to green rosette using mixed infection in the field (GRV + and its sat-RNA + GRAV) and single GRV infection under greenhouse conditions were reported from Nigeria by Olorunju et al. (1992). There was an exception from the RMP12 x M124.781 crosses, where in F2 generation, the plant segregated into 1 susceptible: 3 resistant, sug-gesting dominant gene action governing rosette resistance (Olorunju et al., 1992).Amin (1985) reported a high level of resistance to A. cracivora in some crosses under greenhouse conditions. Progenies of A. chacoense and A. villas interspecific derivatives with cultivated groundnut also showed high resistance to A. crracivora. Resistance to aphid vector identified in cultivated groundnut ICG 5240 [EC36892] (Padgham et al., 1990) was reported to be controlled by single a recessive gene (van de Merwe, 2001; Herselman et al., 2004).
Breeders have largely used the diallel mating scheme to estimate the potential value of genotypes, and their combining ability effects for resistance to foliar disease in groundnut from a fixed or randomly chosen set of parental lines (Adamu et al., 2008). The studies of combining ability provide a guideline for selecting elite parents or combiners which may later be hybridized to accumulate fixable genes through selection. Both SCA and GCA have been reported to be significant in conditioning resistance to foliar disease in groundnut (Vishnuvardhan et al., 2011). Pensuk et al. (2002), from a 6 x 6 diallel cross of resistance to peanut bud necrosis tospovirus (PBNV) reported highly significant GCA effects for PBNV incidence in F2 and F3 generations. SCA was also significant, but the relative contribution to variation among crosses was lesser than those of GCA effects. In an earlier study, Anderson et al. (1990) reported significant GCA and SCA effects for peanut stripe virus (PStV) and rust incidence from a study of diallel in groundnut. Makne (1992) found significant SCA for seed weight per plant, number of pods per plant and pod weight per plant and concluded that these traits were controlled by a non-additive gene action. Adamu et al. (2008) recommended that selection for pod yield and resistance to groundnut rosette disease should be done among progenies from RMP12/ICGV87281 and RMP12/ICGV87018 since they showed best general combiners for these traits. He also suggested that the significance of SCA mean squares for some of the traits is an indication that non-additive gene effects played an important role in their inheritance. SCA mean square was much smaller than GCA mean squares, which indicated that additive genetic variance was more important than non-additive genetic variance for these traits. Studies on combining ability in F2 and F3 crosses of Spanish and Virginia groundnut have shown that GCA and SCA were significant for almost all traits (Ali et al., 2001) with preponderance of SCA which implies that selection for pod yield would be more effective in later generations. However, greater magnitude of GCA effect over SCA has been reported indicating the importance of additive genetic variance over non-additive variance. The mating designs have been used extensively to study the genetics of resistance to viral diseases in wheat, such as wheat soil borne mosaic virus, Barley yellow dwarf virus and wheat streak mosaic virus (Dubey et al., 1970; Cisar et al., 1982; Hakizimana et al., 2004).
From the available reports, it is evident that information on the precise nature of genetic control of GCRD in groundnut is still lacking. Appropriate experimental design that includes the GRD resistant lines should provide additional information on the gene action involved in the expression of resistance. The knowledge on combining ability and type of gene action responsible for regulation of expression of GCRD would certainly help in planning for appropriate breeding strategies. The objective of this study was to determine the mode of inheritance of resistance to GCRD.
MATERIALS AND METHODS
Population development and phenotype evaluation
The study involved the use of nine experimental lines comprising of three aphid resistant (ICGX – SM 00020/5/9, ICGX – SM 00017/5/P10/P1 and ICGX – SM 00020/5/P4/P1 and three GCRD resistant (ICGV IS 07890, ICGV IS 07899 and ICIAR-19BT) genotypes (these genotypes were previously evaluated for three years (2008 – 2010) at the Institute for Agricultural Research (IAR), Ahmadu Bello University (ABU),Zaria, Nigeria and were confirmed to have field resistance to aphids and GCRD)obtained from the International Crop Research Institute for Tropical Agriculture (ICRISAT) inMali. Three widely cultivated varieties (SAMNUT14, KWANKWASO, and MANIPENTA) were also included as parents in the population development. The pedigree descriptions of the nine genotypes are presented in Table 1. The genotypes were manually cross-pollinated in a half diallel mating scheme at the screen house of IAR, SamaruIAR) Samaru, (11°10.00?N and 7°38.00? E, 693 m), and ABU in 2011. Additional manual cross-pollinations were made at IAR during the 2011 rainy season. Seed limitations for multi-location evaluation were overcome by advancing F1 seeds to next generation (F2) as suggested by (Hallauer et al., 2008).
The nine parental lines along with the 36 F2 progenies were evaluated for GCRD resistance using a 9 x 5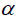 lattice design with two replications at two locations (Samaru, Kaduna state, and Lafia, Nasarawa state (8°32"N, 7°42"E ) during the 2011/2012 growing seasons using an infector – row techniques (susceptible SAMNUT 14 genotype was planted in alternate rows with test materials) as described by Olorunju et al. (2001) at the two locations. The infector rows were planted 2 weeks prior to the test materials to allow the build-up of inoculum. Two row plots of 4.0 m in length with inter and intra-row spacing of 0.75 m x 0.25 m, respectively, were used.
lattice design with two replications at two locations (Samaru, Kaduna state, and Lafia, Nasarawa state (8°32"N, 7°42"E ) during the 2011/2012 growing seasons using an infector – row techniques (susceptible SAMNUT 14 genotype was planted in alternate rows with test materials) as described by Olorunju et al. (2001) at the two locations. The infector rows were planted 2 weeks prior to the test materials to allow the build-up of inoculum. Two row plots of 4.0 m in length with inter and intra-row spacing of 0.75 m x 0.25 m, respectively, were used.
Aphid and GCRD resistance evaluation
A. craccivora colonies were collected from infested cowpea Vigna unguiculata L., and groundnut A hypogaea plants at different locations in groundnut producing area in Nigeria to cover the different isolates that may be present in the country. The colonies (presumed to be viruliferous) were each maintained on susceptible groundnut genotype SAMNUT 14 in a screen house.
Two wingless (apterae) aphids were transferred onto 7 to 14-day-old seedlings of nine parental lines and their 36 F2 progenies grown at IAR. Each genotype was observed for the presence or absence of aphid colonies (adults as well as nymphs) 7 days after infestation. Plants with no aphid colonies were re-infested with aphids seven days after the first infestation. It is rare to find plants without aphids in choice tests because the aphids are free to roam to find suitable plant hosts. Aphids that appeared to be transient, possibly probing for feeding sites, are often observed on resistant plants in choice tests, along with dead aphids. Sometimes several viviparous aptera, surrounded by a few nymphs, were observed on resistant plants without the development of established colonies. Rate of aphid infestation was evaluated using 0-4 scale, combined with GCRD incidencedeveloped by Mensah et al. (2005, 2008). Aphid infestation three time (at forthright interval) by a 0 = No aphid, 0.5 = fewer than 10 aphids per plant, no colony formed, 1.0 =11–100 aphids per plant, plants appear healthy, 1.5 = 101–150 aphids per plant, plants appear healthy, 2.0 = 151–300 aphids per plant, mostly on the young leaves or tender stems, plants appear healthy, 2.5 = 301–500 aphids per plant, plants appear healthy, 3.5 = 501–800 aphids per plant, young leaves and tender stems are covered with aphids, leaves appear slightly curly and shiny, 3.5 = more than 800 aphids per plant, plants appear stunted, leaves appear curled and slightly yellow, no sooty mould and few cast skins and 4.0 = more than 800 aphids per plant, plants appear stunted, leaves appear severely curled and yellow and are covered with sooty mould and cast skins. An aphid damage index (DI) for each line was calculated by the following formula: DI = ∑ (scale value x no. of plants in the category)/ (4 x total no. of plants) x 100. The DI ranges between 0 for no infestation and 100 for the most severe damage (Mensah et al., 2005). The DI was used as an indicator of aphid resistance and was applied in the analysis. The disease severity was recorded as the amount of plant tissue that is diseased, chlorotic rosette.
Reaction to GCRD was recorded on a scale of 1 to 9 as described by GGP (2000) as follows: 1 = No apparent rosette symptoms, 3 = 10 – 20% rosette symptoms, 5 = 20 to 60% rosette symptoms, 7 = 60 – 80% rosette symptoms and 9 = 100% rosette symptoms. The results of these observations were transformed to compute infection responses as measured by area under disease progressive curve (AUDPC) based on Moldovan et al. (2005).
Agronomic data such as pod weight per plant (g), and sound kernel weight per plant (g) were measured on five randomly selected plants per plot.
Data analysis
Genetic analysis of resistance to GCRD
Analysis of the diallel for general combining ability (GCA)andspecific combiningability (SCA) for all traits were based on the Model I, method 2 proposed by Griffing (1956). Parents and one set of F2’s, excluding reciprocal F2’s, were included in the analysis combining abilities.
Trait values were predicted based on traits mean value to produce a balanced data set. Diallel data were analysed using the Diallel SAS-05 program (Zhang et al., 2005). GCA and SCA effects were determined for parents and the 36F2’s, respectively. The following linear mixed model was fitted to data to estimate variance components for single and multi-location diallel tests. The model for the analysis of variance for single location was: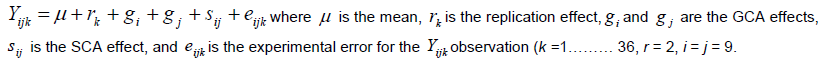
Model for the analysis of variance for multi-location was:
Baker’s ratio
Prediction of progeny performance based on GCA and SCA is carried out by the use of Baker’s ratio, which was the ratio of combining ability variance component described by Baker (1978) as follows: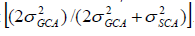 . The closer this ratio was to unity, the greater the predictability based on GCA alone.
. The closer this ratio was to unity, the greater the predictability based on GCA alone.
RESULTS
General and specific combining ability for the traits
For the traits studied, the mean square (MS) values for both GCA and SCA were significant (P < 0.05) (Table 2). The SCA mean squares were higher than the GCA mean squares for DI and AUDPC (Table 2). The magnitude of GCA x location interaction for the majority of traits were relatively small compare to the GCA mean squares. Significant SCA × location interactions (p < 0.05) were observed for all traits. The GCA and SCA variance components were significantly different from zero for all the traits (Table 3). To understand the relative importance of general and specific combining abilities for DI and AUDPC, estimates of components of GCA and SCA that approximates variances estimated according to Bakers ratio (Baker, 1978) indicates a ratio of closer to unity (0.73) for aphid damage index and low value
Partitioning of genotypes into genetic effects indicated significant (p < 0.01) GCA and SCA effects for all the traits. The GCA effects for AUDPC ranged from – 10.61 for ICGX - SM 00020/5/9 to 5.44 in MANIPENTA (Table 5). Genotypes with the lowest desirable negative GCA effects were ICGX – SM 00020/5/9 (-10.61), SAMNUT14 (-2.04), ICGV IS 07890 (-1.58) and ICGX – SM 00017/5/P10/P1 (-1.41).The highest GCA effects for this trait were exhibited by KWANKWASO (5.44) and MANIPENTA (3.58) which were the most susceptible genotypes in this study. The GCA effect for sound kernel weight per plant (g) was highest in ICGV IS 07890 (5.74) and lowest in MANIPENTA (- 9.25). The parents, ICIAR-19BT (4.69), ICGX - SM 00020/5/9 (3.78) and ICGX - SM 00017/5/P10/P1 (3.10) depicted significantly high positive GCA effects.
The specific combining ability effects for AUDPC ranged from -15.65 to 35.96. Most crosses revealed positive SCA effect, 13 out of 36 crosses (36.11 %) had negative SCA effects (Table 5). The F2 combinations, ICGX - SM 00017/5/P10/P1 X ICIAR-19BT (-15.65) had the best desirable negative SCA effects. Other crosses with desirable negative and significant SCA effects for this trait includes ICGX – SM 00020/5/P4/P1 X SAMNUT 14 (-9.66), ICGV IS 07899 X SAMNUT 14 (–8.73) and ICGX - SM 00017/5/P10/P1 X ICGV IS 07890 (–8.1). In contrast, ICGV IS 07890 X ICGV IS 07899 (-7.64), ICGX - SM 00020/5/9 X MANIPENTA (-7.22) and ICGX – SM 00020/5/9 X SAMNUT 14 (-5.52) had high negative but not significant SCA effects. The greatest SCA effect (35.96) was recorded for ICGX - SM 00020/5/P4/P1 X MANIPENTA.
Other crosses depicting significantly positive SCA effects includes ICGX – SM 00020/5/P4/P1XICGV IS 07890 (12.56), ICGV IS 07890XMANIPENTA (15.48), ICGX - SM 00017/5/P10/P1XKWANKWASO (15.84) andICGX - SM 00017/5/P10/P1XSAMNUT 14(18.61) (Table 4).
The SCA effects for sound kernel weight per plant (g) was highest in ICGV IS 07890 X SAMNUT 14 (19.03) and lowest in ICGX – SM 00020/5/P4/P1 XICGV IS 07890 (- 16.07) (data not shown). The other parents that recorded the significantly high SCA effects were ICGV IS 07899XICIAR-19BT (15.17), SAMNUT14 X KWANKWASO (13.94), ICIAR-19BT X SAMNUT14 (11.55), ICGX - SM 00020/5/9 X ICGV IS 07899 (7.88), ICGX - SM 00020/5/9XICGX - SM 00020/5/P4/P1 (7.74), ICGX - SM 00020/5/9XICGV IS 07890 (7.22) and ICGX -SM 00017/5/P10/P1 X ICIAR-19BT (6.75). Whereas ICGX – SM 00020/5/9 X ICGX – SM 00017/5/P10/P1 (-10.64), ICGV IS 07890 X ICGV IS 07899 (-13.84) and ICGX – SM 00020/5/P4/P1 X ICGV IS 07890 (-16.07) had the lowest SCA effects for sound kernel yield per plant (data not shown). However, the F2s that combined significant and desirable SCA effects for SKWPT and AUDPC were ICGX – SM 00017/5/P10/P1XICIAR-19BT (6.75, –15.67), ICGX – SM 00020/5/9XICGX – SM 00020/5/P4/P1 (7.74, –2.77), ICGX – SM 00020/5/P4/P1XSAMNUT 14 (6.00, – 9.66) and ICGV IS 07899XKWANKWASO (3.75, –2.10).
DISCUSSION
Both GCA and SCA made significant and important contribution to progeny variation for DI and AUDPC. All the parents of the most GCRD-resistant crosses; ICGX – SM 00017/5/P10/P1, ICIAR-19BT, ICGX – SM 00020/5/9 and ICGX – SM 00020/5/P4/P1 had appreciable resistance and also favourable GCA resistance values. This suggests that, although resistance to GCRD tends to be at least partly dominant (Clements et al., 2004), optimal resistance in progenies will require crossing parental genotypes that are both GCRD resistant, supporting the findings of Loffler et al. (2011) and Hung and Holland (2012). Therefore, selection for resistance should not be confined to a single group but should be performed in parallel in all groups. The study showed that crosses between two susceptible genotypes resulted in progenies with susceptibility to GCRD. In contrast, Loffler et al. (2011) observed that hybrids often had more disease than their parental inbreds, perhaps because they used highly susceptible tester lines and higher inoculum pressure for hybrids than parents. SCA as well as GCA were important for evaluating resistance of the progeny. Significant SCA detected in 13 of 36 possible combinations indicated the presence of non-additive gene effect. Significant SCA effects were observed for the combinations ICGX – SM 00017/5/P10/P1 X ICIAR-19BT (aphid resistant//rosette resistant), ICGX – SM 00020/5/9 X ICGX – SM 00020/5/P4/P1 (aphid resistant//aphid resistant), ICGX – SM 00020/5/P4/P1 X SAMNUT 14 (aphid resistant//rosette susceptible) and ICGV IS 07899 X KWANKWASO ((aphid resistant//rosette susceptible). These results indicate resistance of these progenies was higher than would be expected from average of their expected parents based on AUDPC symptom rating. The largest positive SCA effects correspond to ICGX – SM 00020/5/P4/P1 X MANIPENTA. This combination was more susceptible than predicted average parent performance indicating the importance of non-additive gene effect in this particular cross. Kenga et al. (2004) suggest that the difficulty in predicting the resistance level of the hybrid, on the basis of GCA alone should necessitate testing of specific male-female combinations. The SCA values provide important information about the performance of the hybrid relative to its parents. Arunga et al. (2010) found that the SCA effect alone has limited value for parental choice in breeding programs. The authors suggested that per se performance of the lines, SCA and GCA effects should be considered in selecting desirable parents in a breeding programme which is desired by any breeder. Furthermore, it was observed that crosses involving one good combiner and one average or poor combiner showed negative SCA effects. For example, MANIPENTA and KWANKWASO had poor GCA values for GRD resistance, while their crosses with ICGV IS 07899 and ICGX – SM 00020/5/9, respectively, had significant and desirable SCA effects. This is in agreement with Habarurema et al. (2012) which made similar conclusion in a study on bacterial blight ((Xanthomonas oryzae pv.oryzae) in rice.
The combining ability ratio, also known as Baker’s ratio, for resistance to GCRD observed in this study was less than unity. According to Baker (1978), when combining ability ratio approaches unity, GCA alone cannot predict the performance of the parents. Thus, the GCA scores could not be used to predict the performance of the parents in the present study, because the value of Baker’s ratio is much lower than the theoretical maximum of unity. Low Baker’s ratio observed for AUDPC in this study highlighted the importance of SCA variance, and hence the importance of dominance and/or epistatic gene effects for increasing resistance to GRD. This implies that selection should be done on latter generations, based on better hybrid combinations, rather than the performance of the parents involved in crossing programs. Partitioning G x L into variance to GCA x L and SCA x L interaction effects indicated significant variances of both GCA x L and SCA x L effects. The significant of GCA x L variance implied that GCRD symptom rating (AUDPC) was sensitive to environmental conditions and data from additional environments or seasons will lead to precise GCA .
CONCLUSIONS AND RECOMMENDATIONS
The estimates of low values of ratio of combining ability variance indicated that nonadditive gene effects were more important than additive gene effects in determining GCRD resistance in groundnut germplasm evaluated in this study. The nature of genetic variation for aphid damage index and AUDPC and its relationships with sound kernel yield implies that planning for successful breeding program for GRD resistance will be possible. ICGX – SM 00017/5/P10/P1, ICIAR-19BT, ICGX – SM 00020/5/9 and ICGX – SM 00020/5/P4/P1 were found to have good GCA for GCRD resistance and could be deployed in groundnut breeding programmes to improve the level resistance to the GCRD. The study recommends multi-location evaluations of advanced breeding lines in a range of environments and also to identify environments that effectively discriminate genotypes based on reaction to GCRD.
CONFLICT OF INTEREST
The authors have not declared any conflict of interest.
REFERENCES
| Adamu AK, Olorunju PE, Ado SG, Alabi SO (2008). General and Specific Combining Ability Estimates for Rosette Resistance, Early Maturity and other Agronomic Traits in Groundnut (Arachis hypogaea L.) Int. J. Pure Appl. Sci. 2(1):33-41. | ||||
| Ali N, Nawaz MS, Bashir K, Mirza MY (2001). Combining Ability estimates in F2 and F3 Generations for Early Maturity and Agronomic Traits in Peanut (Archis yhpogaea L.). Pakistan J. Bot. 33(1):93-99. | ||||
| Anderson WF, Patanothai A, Wynne JC, Gibbons RW (1990). Assessment of a diallel crosses for multiple foliar pest resistance in peanut Olaugineux., 45(8/9): 373-378. | ||||
| Amin PW (1985). Resistance of wildd species of groundnut to insect and mite pests. In Cytogenetics of Arachis. Proceedings of the International Workshop, 31 Oct – 2 November 1983, ICRISATT Center, India. Patancheru, Andhra. Pradesh 502 324, India: International Crops Research Institute f or Semi-Arid Tropics. | ||||
| Arunga EE, Van Rheenen HA, Owuoche JO (2010). Diallel analysis of Snap bean (Phaseolus vulgaris L.) varieties for important traits. Afr. J. Agric. Res. 5(15):1951-1957. | ||||
|
Baker RJ (1978). Issues in diallel analysis. Crop Sci. 18(4):533-536. Crossref |
||||
| Bock KR, Nigam SN (1988). Methodology of groundnut rosette screening and vector-ecology studies in Malawi. In Coordinated research on groundnut rosette virus disease. Patancheru, Andhra Pradesh 502 324, India: International Crops Research Institute for the Semi-Arid Tropics (ICRISAT), pp.6-10. | ||||
|
Bock K, Murant A, Rajeshwari R (1990). The nature of the resistance in groundnut to rosette disease. Ann. Appl. Biol. 117:379-384. Crossref |
||||
| Cisar C, Brown CM Jedlinski H (1982). Diallel analysis for tolerance in winter wheat to barley yellow dwarf virus. Crop Sci. 27:178-180. | ||||
|
Clements MJ, Maragos CM, Pataky JK, White DG (2004). Sources of resistance to fumonisin accumulation in grain and Fusarium ear and kernel rot of corn. Phytopathol. 94:251-260. Crossref |
||||
| Berchoux de CD (1960). La rosette de l.arachide en Haute-Volta. Comportement de lignées résistantes. Oléagineux, 15:229-223. | ||||
|
Dubey SN, Brown CM, Hooker AL (1970). Inheritance of field reaction to soil – borne wheat mosiacvirus. Crop Sci. 10:93-95. Crossref |
||||
| Griffing B (1956). Concept of general and specific combining ability in relation to diallel crossing system. Australian J. Biol. Sci. 9:463-493. | ||||
| Habarurema I, Asea G, Lamo J, Gibson P, Edema R, Séré Y, Onasanya RO (2012). Genetic analysis of resistance to rice bacterial blight In Uganda. Afr. J. Crop Sci. 105-112. | ||||
|
Hakizimana F, Ibrahim AMH, Langham MAC, Haley SD, Rudd JC (2004). Diallel Analysis of Wheat streak mosaic virus Resistance in Winter Wheat. Crop Sci. 44:89-92. Crossref |
||||
| Hallauer AR, Miranda JB, Carena MJ (2008). Quantitative genetics in maize breeding. 3rd ed. Iowa State University Press, Ames, IA. | ||||
| Harkness C (1977). The breeding and selection of groundnut varieties for resistance to rosette virus disease in Nigeria. Report submitted to African Groundnut Council. 45pp. | ||||
|
Herselman L, Thwaites R, Kimmins FM, Courtois B, van der Merwe PJA, Seal SE (2004). Identification and mapping of AFLP markers linked to peanut (Arachis hypogaea L.) resistance to the aphid vector of groundnut rosette disease. Theor. Appl. Gen. 109:1426-1433. Crossref |
||||
| Hung HY, Holland JB (2012). Diallel Analysis of Resistance to Fusarium Ear Rot and Fumonisin Contamination in Maize. Crop Sci. 52:2173–2181. | ||||
|
Janila P, Nigam SN, Pandey MK, Nagesh P, Varshney RK (2013). Groundnut improvement: use of genetic and genomic tools. Front. Plant Sci. 4: 1-16. Crossref |
||||
|
Kenga R, Alabi SO, Gupta SC (2004). Combining ability studies in tropical sorghum (Sorghum bicolor (L.) Moench). Field Crops Res. 88:251-260. Crossref |
||||
|
Loffler M, Kessel B, Ouzunova M, Miedaner T (2011). Covariation between line and testcross performance for reduced mycotoxin concentrations in European maize after silk channel inoculation of two Fusarium species. Theor. Appl. Gen. 122:925-934. Crossref |
||||
| Makne VG (1992). Diallel analysis for studying the inheritance of branches, developed pods and harvest index in groundnut. J. Maharastra Agric. Uni. 17:153-154. | ||||
| Moldovan V, Moldovan M, Kadar R (2005). Assessment of winter wheat cultivars for resistance to Fusarium head blight. Ann. Wheat Newslett. 51:97-98. | ||||
|
Murant AF, Rajeshwari R, Robinson DJ, Raschke JH (1988). A satellite RNA of groundnut rosette virus that is largely responsible for symptoms of groundnut rosette disease. J. Gen. Virol. 69:1479-1486. Crossref |
||||
|
Olorunju PE, Ntare BR, Pande S, Reddy SV (2001). Additional sources of resistance to groundnut rosette disease in groundnut germplasm and breeding lines. Ann. Appl. Biol. 139:259-268. Crossref |
||||
|
Padgham DE, Kimmins FM, Ranga RGV (1990). Resistance in groundnut (Arachis hypogaea L.) to Aphis craccivora Koch. Ann. Appl. Biol. 117:353-358. Crossref |
||||
|
Pensuk V, Wongkaew S, Jogloy S, Patanothai A (2002). Combining ability for resistance in peanut bud necrosis tospovirus (PBNV). Ann. Appl. Biol. 141(2):141- 146. Crossref |
||||
|
Reddy DVR, Murant AF, Duncan GH, Ansa OA, Demski JW, Kuhn CW (1985). Viruses associated with chlorotic rosette and green rosette diseases of groundnut in Nigeria. Ann. Appl. Biol.107:57-64. Crossref |
||||
|
Taliansky ME, Robinson DJ, Murant AF (2000). Groundnut rosette disease virus complex: biology and molecular biology. Adv. Vir. Res. 55:357-400. Crossref |
||||
| Van der Merwe PJA, Subrahmanyam, P, Hildebrand GL, Reddy LJ, Nigam SN, Chiyembekeza, AJ, Busolo-Bulafu, CM, Kapewa T (2001). Registration of groundnut Cultivar ICGV-SM 90704 with Resistance to Groundnut Rosette International Arachis Newsletter [Publication type: JOURNAL] | ||||
| Vishnuvardhan KM, Vasanthi RP, Reddy KH (2011). Combining ability of yield, yield traits and resistance to late leaf spot and rust in groundnut. J. SAT Agric. Res. 9. | ||||
| Yayock JY, Rossel HW, Harkness C (1976). A review of the 1975 groundnut rosette epidemic in Nigeria. Samaru Conference Paper 9. Zaria, Nigeria: Institute for Agricultural Research (Samaru), Ahmadu Bello University, 12. | ||||
|
Zhang Y, Khang MS, Lamkey KR (2005). DIALLEL-SAS05: A comprehensive program for Griffing's and Gardner – Eberhart analyses. Agron. J. 97:1097-1106. Crossref |
||||
Copyright © 2024 Author(s) retain the copyright of this article.
This article is published under the terms of the Creative Commons Attribution License 4.0