ABSTRACT
This study was carried out to analyze the polymorphic region of exon 1 of OsHKT1;5 of 22 high yielding rice varieties that are salt tolerant. The result of the phenotypic screening for salinity stress was done using the standard protocol of the International Rice Research Institute which recorded that all rice varieties developed very well under control condition, 85.38% were quite tolerant to salinity treatment of 4‰, 68.18% were moderately tolerant to salinity treatment of 8‰ and 36.36% were moderately tolerant to salinity treatment of 10‰, similar to Pokalli. The result of sequencing exon 1 region of OsHKT1;5 gene recorded five Single Nucleotide Polymorphisms (SNP) markers. Five nucleotide substitutions in coding sequence of OsHKT1;5 were found at the positions: 382, ​​418, 484, 551 and 994. All five non-synonymous nucleotide substitutions caused changes in amino acids (D51N, P63A, V86I, R107H and H255D).
Key words: High yielding rice, OsHKT1;5, single nucleotide polymorphisms (SNP) makers.
In recent years, the Mekong Delta has been identified as one of the most seriously affected delta by climate change, where salinity intrusion has caused serious influence to agricultural production of the whole region (IPCC, 2007; ADB, 2009). The complicated development of drought and salinity intrusion in early 2016 is considered the most serious in the past 100 years, causing severe influence on rice production in the Mekong Delta provinces (Ministry of Science and Technology, 2016). High salt concentration affects seed germination, plant growth and crop productivity (Hussain et al., 2017), in which, rice is one of the salinity sensitive species (Maas and Hoffman, 1977; Hussain et al., 2017). According to Turan et al. (2012), in salinity environment, excess Na+ leads to the loss of ionic homeostasis. Potassium acts as a coenzyme for many cytoplasmic enzymes, but when excess Na+ is present in rhizosphere, it competes for K+ particularly at low affinity K+ channels, leading to low K+/Na+ ratio in cytoplasm. Genetic engineering of genes for antiporter or ion channels have been successful in generation of salt tolerant plants by maintaining higher K+/Na+ ratio. In addition, Na+ in reproductive organs prevents photosynthesis and transports starch, therefore reducing rice yields (Hussain et al., 2017).
The capability of salinity tolerance in rice has been reported as an extreme complex mechanism controlled by many genes (Reddy et al., 2017). Many plant membrane transport channels play a major role in stress resisting mechanisms, especially Na+ and K+ channels related to salinity tolerance (Hamamoto et al., 2015). Studies on ion balance in plants have shown that protein HKT (High-Affnity K + Transporter) plays an important role in ion balance in the cells. This gene family includes many genes such as: OsHKT1;2, OsHKT2;3, OsHKT1;3, OsHKT1;1, OsHKT1;4, OsHKT2;1, and OsHKT1;5 (Waters et al., 2013; Hamamoto et al., 2015). In particular, OsHKT1; 5 plays the role of encoding ion Na+ transport proteins from roots to shoots and control shoot Na+concentration in rice (Ren et al., 2005) as the key mechanism to salinity tolerance of rice. In another study, a novel function of OsHKT1;5 was reported in mediating Na+ exclusion in the phloem to prevent Na+ transfer to young leaf blades (Kobayashi et al., 2017). The OsHKT1;5 is about 4,487 bp length in which coding region takes 1,665 bp, consisting of 3 exon regions with their length of 1,235 bp for exon 1, 231 bp for exon 2, and 199 bp for exon 3 (www.rapdb.dna.affrc.go.jp). Therefore, the exon 1 was advantagious for sequencing and also containing enough information to detect SNPs. This study focused on polymorphic sequence analysis of exon 1 of gene OsHKT1;5 in high yielding rices showing changes in protein structure and finding out the role of salt tolerance mechanism in helping breeding of rice varieties in the current period.
Rice materials
Seeds of 22 rice cultivars were provided by The Cuu Long Delta Rice Research Institute (CanTho, Vietnam), including 20 high yielding rice cultivars, Pokkali was used as salinity tolerant control and IR28 used as salinity sensitive control.
Phenotypic screening for salinity stress
In this study, 22 rice cultivars were used for salinity screening at the seedling stage following International Rice Research Institute (IRRI) standard protocol using Yoshida nutrient solution (Yoshida et al., 1976). The seedlings allowed to grow to a height of 1.5 to 2.0 cm, were placed in the boxes on the meshed foam sheet and then placed in a rectangular plastic container filled with 10 L of Yoshida nutrient solution. After 3 days, removed the weakest seedling, keeping three seedlings per hole. The experiment was arranged in a completely randomized design (CRD) with 3 replications at four NaCl concentrations of 0, 4, 8 and 10‰ (1 M of NaCl containing 58.44 g/L, while 4, 8, and 10‰ consisting of 4, 8, and 10 g/L NaCl, so they become 68, 136, and 171 mM, respectively). Nutrient solution was replaced at every 8 day intervals and the pH was maintained at 5 daily. The modified standard evaluation score (SES) of IRRI was used to evaluate the visual symptoms of salt toxicity when IR28 cultivar is completely dead (Table 1).

DNA extraction
Young leaves of 10-days old seedlings of rice cultivars (15 to 20 cm) were used for genomic DNA extraction using the cetyltrimethylammonium bromide (CTAB) method (Rogers and Bendich, 1988).
Primer designing and amplification of OsHKT1;5
The exon 1 region of gene OsHKT1;5 in Nipponbare cultivar was downloaded from the Rice Anotation Project website (www.rapdb.dna.affrc.go.jp), and used to design primer pairs by Primer3Plus software. Total genomic DNA of 22 genotypes was used as template for PCR using the designed primer pairs, including F-5’GGACCTGATCTTCACGTCGG3’ and R-5’GAGCACCATCTCACCGGAG3’. Amplifier product length was 1000 bp.
The mixture for PCR reaction consisted of genomic DNA 50 ng, 1X master PCR buffer kit containing dNTPs and MgCl2, 0.3 µM primers, 1 unit of Taq Polymerase. The PCR was performed in thermocycle with the following steps: 95°C for 2 min of pre-denaturation, 30 cycles (94°C for 30 s, Tm 58°C for 30 s, 72°C for 45 s) and 72°C for 5 min of prolonged extension step. PCR product was run on 2% agarose gel electrophoresis in TBE buffer 1X for 45 min under 140 V. DNA bands were visualized by safe view staining and took gel pictures with Biorad UV gel camera. PCR products that showed only one bright single band on gel were sequenced. Sequencing was done by PhuSa Biochemical Co., Ltd, Can Tho City.
Sequencing the whole exon 1 of OsHKT1;5
From the OsHKT1;5 sequence data of rice cultivars, gene sequences were analyzed using Bioedit for SNPs detection. From SNPs, amino acid sequences in the corresponding protein were inferred and the differences compared among rice cultivars.
Phenotypic screening of rice cultivars at seedling stage for salinity tolerance
Twenty two (22) cultivars of rice seedling were used for screening salinity tolerance. The result recorded in Table 2 and Figure 1 shows that all the rice varieties grew very well in the control condition (0‰) and were also quite tolerant to salinity treatment at 4‰ (19/22 cultivars accounted for 85.38% of the total of cultivars). Pokkali, OM20, OM396, OM429, OM6976, OM5629, OM8108, OM10252, OM355, OM2514, OM9921, OM2517, OM11735, OM8959, and OM6677 were found to be moderately tolerant to salinity treatment of 8‰ (15/22 cultivars accounted for 68.18% of the total of cultivars). There were seven cultivars, namely, OM20, OM396, OM429, OM6976, OM5629, OM8108 and OM10252 that were moderately tolerant similar to Pokalli in salinity treatment of 10‰ (accounted for 36.36% of the total of cultivars); there were three cultivars namely OM6677, OM18 and OM6162 highly susceptible to salinity treatment of 10‰ (accounted for 18.18% of the total of cultivars), similar to IR28.
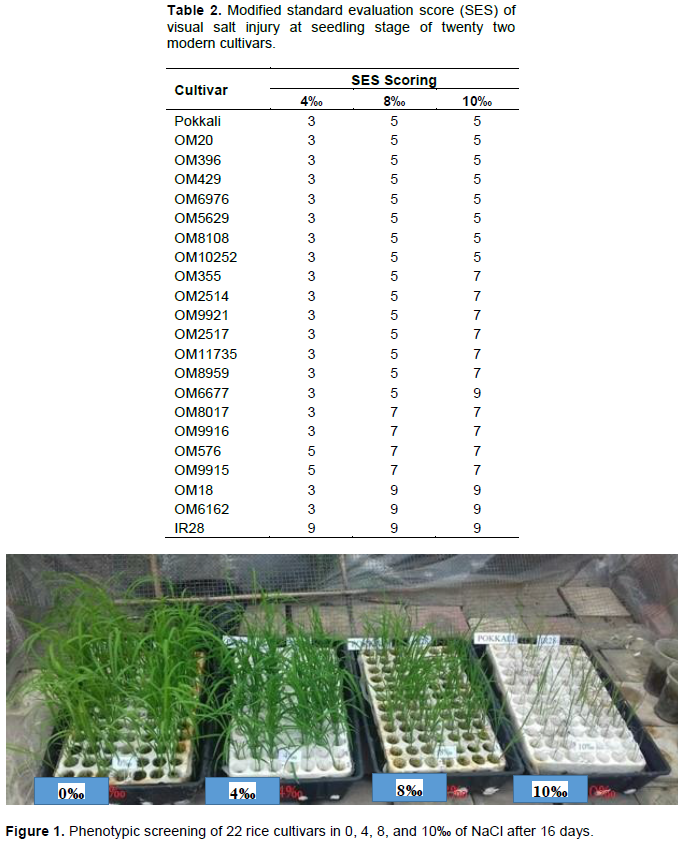
This result was similar to that of Quynh-Hoa et al. (2016) where they recorded that OM6677 was highly susceptible and also similar to that of Nguyen (2012), whereby testing cultivars of AS996, ST20, IR50404, OM6677, and OM6377, they showed that OM6976 developed very well under salt stress among the rice cultivars. Besides, through salinity treatments of 4, 8, and 10‰, the salinity damage increased gradually through phenotypic observation in the surveyed rice varieties.
Analysis of exon 1 region of OsHKT1;5 gene in rice cultivars
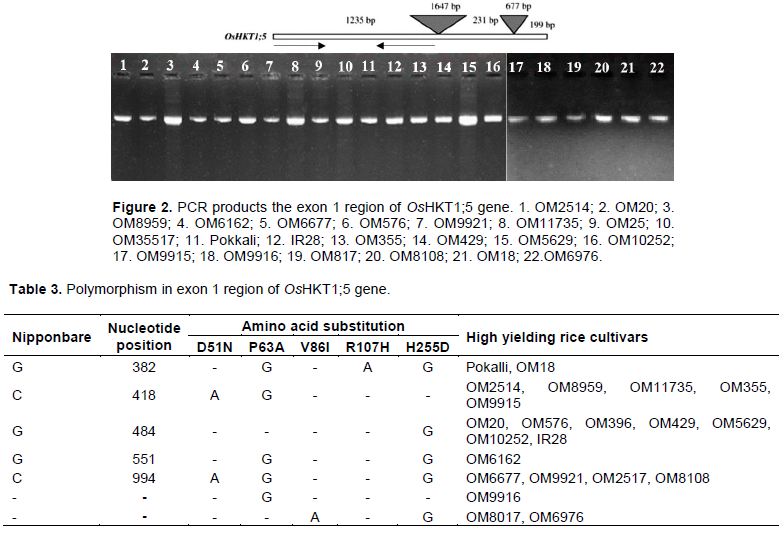
The leaves of 10 to 12 days rice seedlings were collected
for genomic DNA extraction. The extracted DNA was used as template for the amplification of the exon 1 region of OsHKT1;5 gene in the PCR assay. As shown in Figure 2, the PCR products showed only one bright single band on gel, which was specific and with the same size across the rice cultivars, without unintended bands. Thus, the exon 1 region of OsHKT1;5 gene was successfully amplified in all the investigated rice cultivars.
Amplified DNA fragments were purified and the exon 1 region of OsHKT1;5 gene of 22 high yielding rice cultivars sequenced in this research. The stable signal area was recorded as 938 nucleotides from nucleotide 230 to nucleotide 1168. This nucleotide region is used to analyze SNP markers. The result of sequencing exon 1 region of OsHKT1;5 allowed the detection of five nucleotide substitutions at positions: 382, 418, 484, 551, and 994 when compared with the reference sequence in the database (Nipponbare allele). Among the twenty two high yielding rice cultivars, six cultivars recorded three SNP markers. Among these six cultivars, Pokalli and OM18 have three SNP markers at position C418G (C is replaced by G at nucleotide position 418), G551A, C994G; OM6677, OM9921, OM2517, and OM8108 have three same SNP markers at position G382A, C418G, and C994G. Seven cultivars that recorded one SNP marker at position C994G were OM20, OM576, OM396, OM429, OM5629, OM10252, and IR28. Interestingly, IR28 and Nipponbare were used as controls for sensitive cultivars (Table 3).
All the five nucleotide substitutions were non-synonymous substitutions. Five non-synonymous substitutions mean that nucleotide substitutions lead to amino acid substitutions were D51N (Aspartic acid was replaced by Asparagine at position 51), P63A (Proline was replaced by Alanine at position 63), V86I (Valine was replaced by Isoleucine at position 86), R107H (Arginine was replaced by Histidine at position 107), and H255D (Histidine was replaced by Aspartic acid at position 255). It can be seen, in this research, that nucleotide substitutions in the exon 1 region of OsHKT1;5 gene lead to amino acid substitutions in twenty two rice cultivars when compared with Nipponbare cultivar.
Some studies about SNPs of OsHKT1;5 gene were reported: Mishra et al. (2016) carried out the experiment on 299 wild rice accessions collected from different agro-climatic regions of India under salt stress condition. Of these, 95 representative accessions were sequenced for members of HKT ion transporter family genes (the whole OsHKT1;5 gene). The results for OsHKT1;5 gene has 45 NSPs (8 from coding and 37 from non-coding). In addition, the OsHKT1;5 gene of Godawee (a Sri Lankan traditional rice variety known for its salinity tolerance) was sequenced, and 122 SNPs were found (Sanjeewa et al., 2017). In the current study, five SNPs were found in exon 1 of OsHKT1;5 gene leading to five amino acid substitutions of D51N, P63A, V86I, R107H and H255D. It seems to be less polymorphism, but in coding region only. Similarly, Ren et al. (2005) found six SNPs in the OsHKT1;5 coding region that led to four amino acid changes, namely P140A, R184H, H332D and L395V. Negrão et al. (2013) found three nonsynonymous SNPs in OsHKT1;5 coding region leading to three amino acids change consinting of T67K, P140A and R184H, in which two residue differences between ‘Nipponbare’ and IR29 (includes IR64) were observed, speciï¬cally D129N and P140A.
With the present data, as reported by Quynh-Hoa et al. (2016) and Do et al. (2016), it is quite difficult to point out the relationship between nucleotide polymorphism in the exon 1 region of OsHKT1 gene and the salt tolerance level of the testing rice cultivars. It might be helpful to explore the nucleotide polymorphism in OsHKT1;5 gene, which plays role in the regulation of gene expression.
The results of the phenotypic screening for salinity stress, among 22 high yielding rice varieties, recorded that nineteen cultivars were quite tolerant in salinity treatment at 4‰, fifteen cultivars were moderately tolerant in salinity treatment of 8‰ and only eight cultivars were moderately tolerant similar to Pokalli in salinity treatment of 10‰. Besides, through salinity treatments of 4, 8, and 10‰, the salinity damage increased gradually through phenotypic observation in the surveyed rice varieties. The exon 1 region of OsHKT1;5 gene was successfully amplified in all the investigated rice cultivars. The results of sequencing exon 1 region of OsHKT1;5 gene recorded five SNP markers at positions: 382, 418, 484, 551, 994. All five non-synonymous nucleotide substitutions caused changes in amino acids (D51N, P63A, V86I, R107H, and H255D).
The authors have not declared any conflict of interests.
This research was financially supported by Can Tho University, Viet Nam.
REFERENCES
|
Asian Development Bank (ADB) (2009). Socialist Republic of Viet Nam: Climate Change Impact and Adaptation Study in the Mekong Delta. Technical Assistance Report. Project number:43295.
|
|
|
|
Do TP, Nguyen VM, Hoang HY (2016). Assessment of natural variation in OsHKT1;2 gene in rice (Oryza sativa). Vietnam National University Journal of Science: Natural Sciences and Technology 32:189-193.
|
|
|
|
|
Hamamoto S, Horie T, Hauser F, Deinlein U, Schroeder JI, Uozumi N (2015). HKT transporters mediate salt stress resistance in plants: from structure and function to the field. Current Opinion in Biotechnology 32:113-120.
Crossref
|
|
|
|
|
Hussain S, Zhang JH, Zhong C, Zhu LF, Cao XC, Yu SM, Bohr JA, Hu JJ, Jin QY (2017). Effects of salt stress on rice growth, development characteristics, and the regulating ways: A review. Journal of Integrative Agriculture 16(11):2357-2374.
Crossref
|
|
|
|
|
Intergovernmental Panel on Climate Change (IPCC) (2007). Climate Change 2007: The Physical Science Basis. Contribution of Working Group I to the Fourth Assessment Report of the Intergovernmental Panel on Climate Change. Solomon S. Cambridge University Press. Cambridge, United Kingdom and New York, NY, USA.
|
|
|
|
|
Kobayashi NI, Yamaji Naoki, Yamamoto Hiroki, Okubo Kaoru, Ueno Hiroki, Costa Alex, Tanoi Keitaro, Matsumura H, Kashino M. F., Horiuchi T, Nayef Mohammad Al, Shabala S, An G, Ma JF, Horie T (2017a). OsHKT1;5 mediates Na+ exclusion in the vasculature to protect leaf blades and reproductive tissues from salt toxicity in rice. The Plant Journal 91:657-670.
Crossref
|
|
|
|
|
Maas EV, Hoffman GJ (1977). Crop salt tolerance-current assessment. Journal of the Irrigation and Drainage Division 103(2):115-34..
|
|
|
|
|
Ministry of Science and Technology (2016). Saline intrusion in the Mekong Delta - effective solutions to respond to climate change conditions. Access on October 19, 2018. https://www.most.gov.vn (In Vietnamese).
|
|
|
|
|
Mishra S, Singh B, Panda K, Singh BP, Singh N, Misra P, Rai V, Singh NK (2016). Association of SNP haplotypes of HKT family genes with salt tolerance in indian wild rice germplasm. Rice 9:15.
Crossref
|
|
|
|
|
Negrão S, Cecília Almadanim M, Pires IS, Abreu IA, Maroco J, Courtois B, Gregorio GB, McNally KL, Margarida Oliveira M (2013). New allelic variants found in key rice salt-tolerance genes: an association study. Plant Biotechnology Journal 11:87-100.
Crossref
|
|
|
|
|
Nguyen TTT (2012). Selection and regeneration of some rice varieties that are saline tolerant to climate change in the Mekong Delta. Master of Science Thesis, Can Tho University (In Vietnamese).
|
|
|
|
|
Quynh-Hoa P, Xuan-An T, Thi-Nha-Trang N, Thi-Thuy-Anh T, Hai-Yen H, Thi-Hong-Van N, Thi-Hanh T, Thi-Phuc DO (2016). Investigation of polymorphisms in coding region of OsHKT1 in relation to salinity in rice. Rice Science 23(6): 334-338
Crossref
|
|
|
|
|
Reddy IN, Kim BK, Yoon IS, Kim KH, Kwon TR (2017). Salt tolerance in rice: Focus on mechanisms and approaches. Rice Science 24(3):123-144.
Crossref
|
|
|
|
|
Ren ZH, Gao JP, Li LG, Cai XL, Huang W, Chao DY, Zhu MZ, Wang ZY, Luan S, Lin HX (2005). A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nature Genetics 37:1141-1146.
Crossref
|
|
|
|
|
Rogers SO, Bendich AJ (1988). Extraction of DNA from plant tissues. Plant Molecular Biology Manual 6:73-83.
Crossref
|
|
|
|
|
Turan S, Cornish K, Kumar S (2012). Review article Salinity tolerance in plants: Breeding and genetic engineering. Australian Journal of Crop Science 6(9):1337-1348.
|
|
|
|
|
Waters S, Gilliham M, Hrmova M (2013). Plant high-affinity potassium (HKT) transporters involved in salinity tolerance: Structural insights to probe differences in ion selectivity. International Journal of Molecular Sciences 14:7660-7680.
Crossref
|
|
|
|
|
Yoshida S, Forno A, Cock JH, Gomez KA (1976). Laboratory Manual for Physiological Studies of Rice. The International Rice Research Institute. Los Banos.
|
|