ABSTRACT
The effect of drought stress on growth and water relations of four Hibiscus genotypes was investigated. Drought stress significantly retarded vegetative growth as analyzed by plant height, stem diameter, leaf area, fresh and dry weight. The osmotic potential (ys) values decreased with increasing drought time, and the decrease was more pronounced in Fantasia followed by Jay´s orange. Osmotic adjustment (O.A.) increased with drought time, and was greater in Fantasia followed by Jay´s orange and lowest in Annie wood. The genetic diversity among the four hibiscus genotypes was detected by Random Amplified Polymorphic DNA (RAPD) analysis. The data indicate that, thirty-one out of fifty four RAPD markers detected were polymorphic (57.4%). The RAPD specific markers for each genotype were determined. Ten out of the thirty one polymorphic RAPD markers generated were found to be genotype- specific (32.2%). In the meantime, the largest number of RAPD genotype-specific markers was generated for primers OPE-D-07(six markers) while for both of the OPE-B-17 and OP D-05 (two markers each). These markers can be verified as being RAPD markers associated with drought tolerance in the hibiscus genotypes that could help in marker-assisted selection breeding program.
Key words: Hibiscus, biomass production, osmotic adjustment, molecular markers, genetic polymorphism.
Drought is the most important limiting factor for crop production, and it is becoming an increasingly severe problem in many regions of the world (Passioura, 1996; Passioura, 2007; Amin and Hajmohammadrezaei, 2011). Generally, drought stress occurs when the available water in the soil is reduced and atmospheric conditions cause continuous loss of water by transpiration or evaporation (Khaje et al., 2003). Metabolic responses of higher plants to abiotic stresses are complex, since many processes such as carbon metabolism, accumulation of compatible osmolytes, ion partitioning, energy metabolism and growth are modified (Cushman et al., 1990).
Genetic variability within cultivated species originated from closely related ancestors is limited, particularly for traits such as tolerance to environmental stresses (Ch0abane and Valkoun, 1997). Different approaches were used to assay genetic diversity in crop plants including morphological traits, and isozyme electro-phoresis; however these techniques are insufficient to serve as accurate markers due to environmental influences on morphological traits and insufficient polymorphism produced by isozyme approach among closely related genotypes (Yang and Quiros, 1993).
Molecular markers have been developed to give high degree of polymorphism and stability, and provide easier and faster approaches for exploring genetic variability in crop plants (Yang and Quiros, 1993). A molecular marker is a readily detectable sequence of DNA or protein whose inheritance can be monitored. It is the variation in, or polymorphism of, molecular markers, which can be used in genetic diversity studies (Weising et al., 1995).
Random amplified polymorphic DNA (RAPD) developed by Welsh and McCelland (1990) and Williams et al. (1990) proved to be a powerful tool in different genetic analysis. This approach detects DNA polymorphisms based on amplification using a single primer of arbitrary nucleotide sequence of genomic DNA fragments. RAPD markers are attractive because they are simple and quick, very small (nanograms) quantities of DNA are required, automation is feasible, there is no requirement for previous DNA sequence information (Williams et al., 1990), modest cost and ability to detect relatively small amounts of genetic variation (Ragot and Hoisington, 1993).
According to Prasad (2014), RAPD analysis in combination with morphological characters can be used in the identification and determination of the genetic variation between the different varieties and species of Hibiscus. Daudu et al. (2016) assessed the genetic diversity among Nigerian Roselle (Hibiscus sabdariffa Linn.) using certain genetic and morphological markers to group the various accessions collected into distinct genotypes, and they concluded that RAPD-PCR analysis provided very valuable means for determining relationships among H. sabdariffa accessions.
In the present study, four hibiscus genotypes were subjected to drought stress, whiles plant water relation and biomass production were determined. The genetic diversity among these genotypes was resolved using the random amplified polymorphic DNA (RAPD) markers, and thereby the genotype-specific markers for each genotype were determined.
Plant materials and treatments
This study was carried out at the Nursery of the Ornamental Horticulture Department, Faculty of Agriculture, Cairo University, during two winter seasons; 2013 to 2014.The nursery work began on the 1st of June of both seasons. Seedlings from four Hibiscus rosasinensis genotypes namely: Annie wood (Yellow flowers), Cherie(Orange flowers), Jay´s orange(Red flowers) and Fantasia (Pink Flowers) were sown in plastic pots, 25 cm diameter, filled with sandy loam soil prepared specifically for this purpose. Four weeks to post-transplantation, the plants irrigated by tap water were done every 2, 4 and 6 days at a rate of 200 ml/pot. Under each of different period of irrigation, the plants were split into 4 groups; one red, the second pink then orange and the last group was yellow.
The seedlings were harvested at 90 days post treatment. Each watering treatment was replicated four times in a completely randomized design, each replicate contains 3 plants.
Growth parameters
Data on plant height (cm), root length (cm), number of branches/plant, and total dry weights (g)/plant were recorded. Leaf area (cm2) was measured with area meter.
Chemical analysis
Total chlorophyll content (SPAD) determined by using fresh leaf samples were described by Nettoet al. (2005). The proline content (μ moles/g fresh matter) in fresh leaves was also determined according to Bates et al. (1973).
Determination of leaf water relations
The leaf samples were frozen in liquid nitrogen and stored at -20°C. Leaf tissues were thawed and centrifuged at 1,200x g for 25 min at 4°C to extract the cell sap. The osmotic potential (ψs) of the cell sap was measured using a vapor pressure osmometer (model 5,500; Wescor, Logan, UT USA). Osmotic adjustment (OA) was calculated as the difference in ψs between treated and control plants.
Statistical analysis
Statistical analysis was performed using Analyze-it software (Analyze-it, Leeds, UK) in accordance with the method of Maxwell and Delany (1989).
Genetic diversity among hibiscus genotype
DNA extraction and RAPD analysis
Total genomic DNA was isolated using the method descried in Rogers and Bendich (1985). PCR reactions were conducted using arbitrary 10-mer primers (Operon Technology, Inc., Alameda, CA, USA). Six primers out of fifteen RAPD primers tested resulted in clear and repeatable bands. The names and sequences of the primers used are presented in Table 1.
Reaction mixture and PCR condition
The reaction mixture (20 ml) contained 10 ng DNA, 200 mM dNTPs, 1 mM primer, 0.5 units of Red Hot Taqpolymerase (AB-gene Housse, UK) and 10-X Taq polymerase buffer (AB-gene Housse, UK). Samples were heated to 94°C for 5 min, and then subjected to 35 cycles of 1 min at 94°C; 1 min at 35°C and 1 min at 72°C. The amplification products were separated in 1% (w/v) agarose gel in 1 x TBE buffer and visualized by staining withethidium bromide. Reproducibility of DNA profiles was determined by replicating all RAPD reactions at least three times.
Data analysis
All the genotypes were scored for the presence and absence of the RAPD bands. The data were entered into a binary matrix as discrete variables, 1 for presence and 0 for absence of the character, and this data matrix was subjected to further analysis. The Excel file containing the binary data was imported into NT Edit of NTSYS-pc 2.02J. The 0/1 matrix was used to calculate similarity as DICE coefficient using SIMQUAL subroutine in SIMILARITY routine. The resultant similarity matrix was employed to construct dendrogram using sequential agglomerative hierarchical nesting (SAHN), Unweighted pair group method with arithmetic means (UPGMA) to infer genetic relationships and phylogeny. Variations among hibiscus genotypes across the primers were evaluated from pair-wise comparisons for the proportion of shared bands amplified according to Nei (1978).
Plant productivity is greatly affected by drought stress. Availability of water is one of the most important factors, which determine geographical distribution and productivity of plants (
Bartels and
Salamini, 2001). Water stress is perceived as water deficit and can occur with different severity (Ramanjulu and Bartels, 2002). A continuation of a mild water deficit leads to drought and even desiccation (loss of most of the protoplasmic free or bulk water). The response and adaptation of plants to such conditions are very complex and highly variable. It is well documented that, when plant tissues are subjected to drought stress, some physiological and biochemical changes occur. Some of these changes include: accumulation of ABA, closing stomata and reducing leaf area (Premachandra et al., 1995).
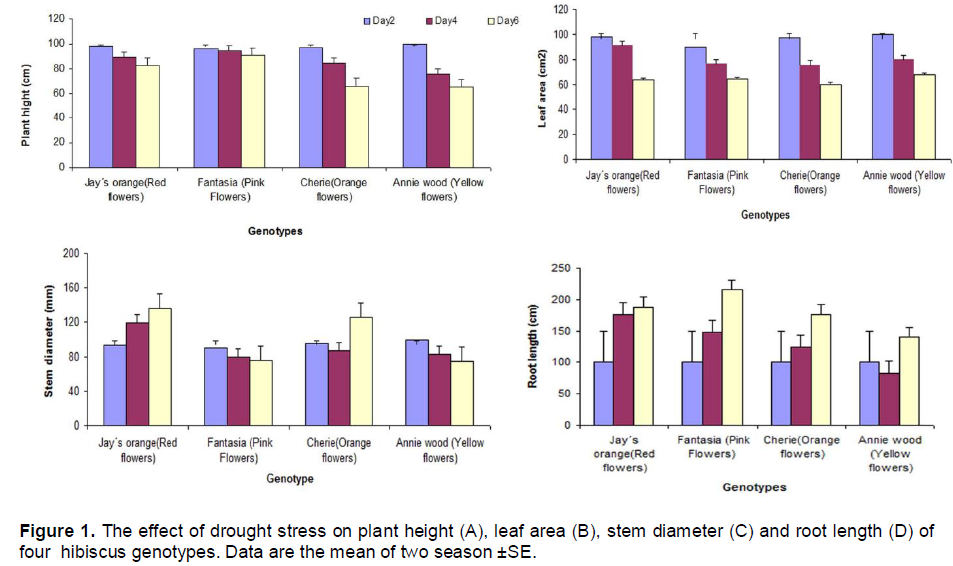
In the present study, four hibiscus genotype namely Annie wood (Yellow flowers), Cherie (Orange flowers), Jay´s orange (Red flowers) and Fantasia (Pink Flowers) were subjected to three water regime (2, 4 and 6 days). The data indicated that the plant height was reduced under drought stress condition in all hibiscus genotypes. The reduction was less in c.v fantasia (pink flowers) while the reduction in plant height was more pronounced in the c.v Annie wood (Yellow flower) six days post-treatment compared to the other genotypes (Figure 1A). The data indicated that drought stress reduced leaf area in all hibiscus genotypes (Figure 1B). Stem diameter increase with increasing drought stress in Jay´s orange and Cherie while the stem diameter of both of Annie wood and Fantasia was reduced with increasing drought time (Figure 1C). Under drought stress, all of the genotypes tested tended to increase their root length in order to find water supply. Fantasia showed the highest root length under long period of drought (six days) compared to the other genotypes (Figure1D). The data presented in Figure 2 indicate that, the biomass production was higher in Cherie which showed higher dry weight compared with the other genotypes (Figure 2).
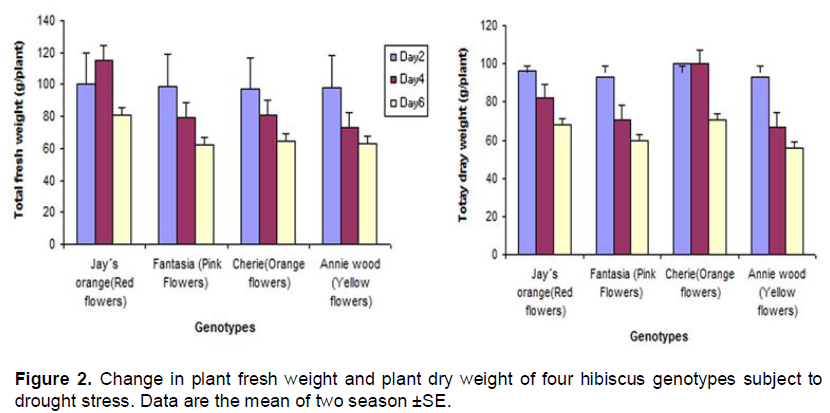
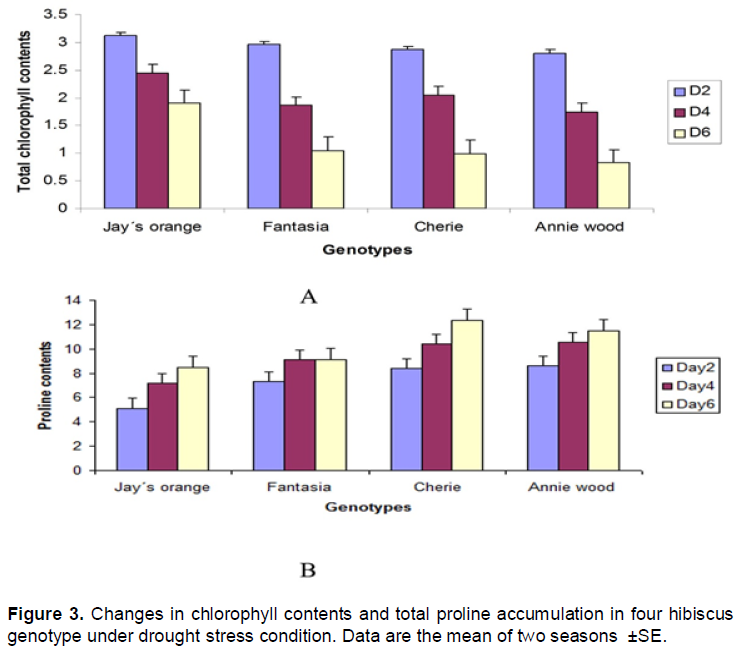
The data indicate that total chlorophyll was reduced by increasing drought time in all genotypes tested (Figure 3A). The reduction in chlorophyll contents results in lowering biomass production as found in Figure 2. It is well known that proline plays a role as an osmo-regulator in abiotic stress tolerance mechanism in plants (Bates et al., 1973). Larcher (2001) indicated that the plantproline concentration increase under drought stress. In the present investigation under drought stress hibiscus plants tends to accumulate higher level of proline which was evident in the present study. Cherie accumulated much higher proline contents followed by Annie wood genotype compared to other genotypes (Figure 3B).
The analysis of variance indicates that the difference between genotypes were significant for plant height, leaf area, fresh weight and dry weight and it was non-significant for stem diameter and root length (Table 2). The data indicate that drought treatment significantly affect the entire earlier mentioned trait except for stem diameter which was non- significant (Table 2).
It is well known that osmotic adjustment involves the net accumulation of solutes in a cell in response to drought stress, and consequently, the osmotic potential decreases, which in turn attracts water into the cells and enables the turgor to be maintained (Neuman et al., 1988).
In the present study, all the parameters of leaf water relations decreased with increasing drought time (Table 3). The osmotic potential (ys) of drought treated plants decreased with increasing of the irrigation time, and the decrease was more pronounced in Fantasia and Jay´s orange genotypes. Osmotic adjustment (O.A.) increased with drought time, and was greater in Fantasia (0.88) followed by Jay´s orange (0.63) and lowest in Annie wood (0.21) (Table 3). The difference in the ability to maintain the osmotic potential under drought stress conditions between the hibiscus genotypes used in the present study reflects the differences in their genetic backgrounds.
Genetic diversity among hibiscus genotypes used
Molecular markers have proved to be useful, both in the identification of individual varieties and in the development of phylogenetic relationships among plant species. Molecular markers have to be developed for each species, from genomic regions that have undergone sufficiently rapid evolutionary change, in order to be useful for discriminating among closely-related germplasm (Cheng et al., 2002; Zehdi et al., 2004; Prasad, 2014; Daudu et al., 2016).
In the present study, in order to investigate the genetic differences among the four hibiscus genotypes used, the random amplified polymorphic DNA (RAPD) analysis was performed. All primers used in the present study resulted in the appearance of PCR products with a variable number of bands. This study shows that a total of 54 DNA markers were detected among the four hibiscus genotypes of which, 31 bands were polymorphic (57.4%) and can be considered as useful RAPD markers for the four hibiscus genotypes used (Figure 4 and Table 4 ). The largest number of RAPD bands was detected for primers OPE-D-07 (12 bands) followed by the primers OPE-B-11 and OPE-B-17 (10 bands each), while the lowest was scored for OPE-F-12 (6 bands).
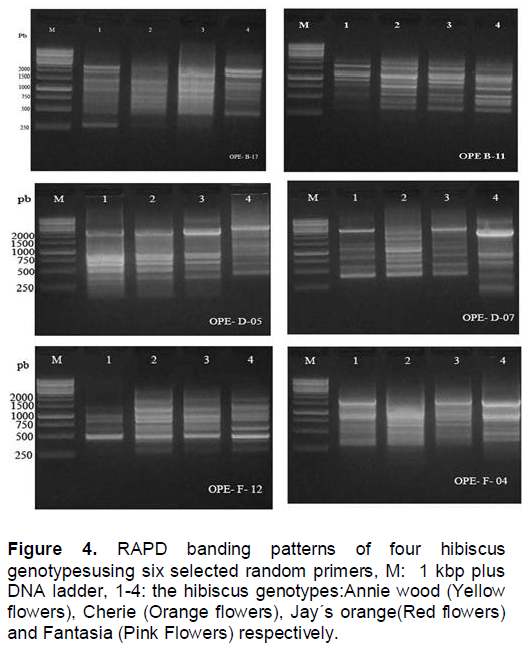
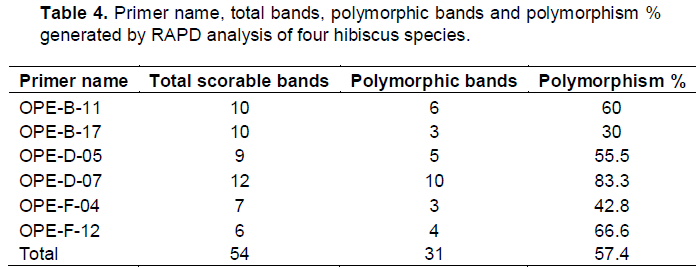
Levi and Rowland (1997) could identify blue berry cultivars, and evaluate their genetic relationship using randomly amplified polymorphic DNA (RAPD) which agreed with the present data, and also support the results of Degani et el. (2001) who estimated the genetic similarities among apple cultivars by RAPD and AFLP analyses. The data presented in Table 4 shows the polymorphic bands generated from each primer.
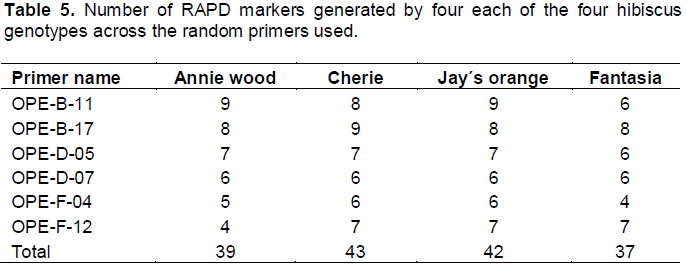
The data presented in Table 5 show that the Cherie (Orange flowers) showed the highest number of RAPD markers (43 markers) followed by Jay´s orange (Red flowers) (42 markers), then Annie wood (Yellow flowers) which recorded 39 markers while Fantasia (Pink Flowers) recorded 37 marker. These results agreed with the data presented by both of Ahmed (2013) and also with Seeruttum and Sanmukiya (2013). The present study is in perfect agreement with the work of Omalsaad et al. (2014), Daudu et al. (2015) and Daudu et al. (2016) on H. sabdariffa. It is also evident from the present results that RAPD molecular marker was able to detect small genetic differences among the different genotypes of Hibiscus rosasinensis.
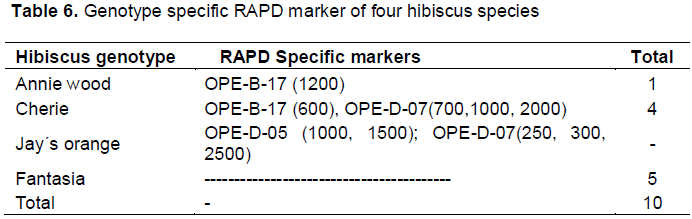
The specific RAPD markers for the different hibiscus genotypes used in the present study are listed in Table 6. Ten out of the thirty one polymorphic RAPD markers generated were found to be genotype- specific (32.2%). The largest numbers of RAPD specific markers were scored for Fantasia (5 markers) and Cherie (4 markers while Annie wood scored only one markers. In the meantime, the largest number of RAPD genotype-specific markers was generated for primers OPE-D-07(six markers) while both of the OPE-B-17 and OP D-05 produced two markers each. From these results, one may conclude that these RAPD-bands can be considered as genetic markers for detecting drought tolerance in the four hibiscus genotypes used.
The RAPD-based phenogram (Figure 5) grouped the investigated genotypes into two main clusters. The first cluster included Fantasia, the second cluster was divided into two sub-clusters, the first sub-cluster included Cherie and the second sub-cluster included Annie wood and Jay´s orange. This may indicate that the Annie wood and Jay´s orange are genetically very close.
Ranking genotypes based on growth rate, physiological parameter including water relation and proline contents it could be suggested that the hibiscus genotype Fantasia and Jay´s orange proved to be drought tolerant. The higher drought tolerance in these cultivars was due to their ability to maintain higher osmotic potential by accumulating much higher concentration of osmoticum solutes. According to the genotype specific molecular markers, these markers can be considered useful for drought tolerance in hibiscus breeding programs.
The authors have not declared any conflict of interests.
The authors acknowledge the Genetic Engineering Research Center staff at Cairo University for their assistance during the molecular markers experiment, also we acknowledge the Department of Ornamental Horticulture team members, for their assistance during field experiment.
REFERENCES
|
Ahmed FAK (2013). Molecular Genetic Identification of Some Egyptian Hibiscus Samples. J. Am. Sci. 9(10):28-35.
|
|
|
|
Amin B, Hajmohammadrezaei M (2011). Effect of drought stress and its interaction with ascorbate and salicylic acid on okra (Hibiscus esculents L.) germination and seedling growth. J. Stress Physiol. Biochem. 7(1):55-65.
|
|
|
|
|
Bates L, Waldern RP, Teare ID (1973). Rapid determination of free Praline for water stress. Stud. Plant Soil. pp. 205-207.
|
|
|
|
|
Bartels D, Salamini F (2001). Desiccation Tolerance in the Resurrection PlantCraterostigmaplantagineum. A Contribution to the Study of Drought Tolerance at the Molecular Level. Plant Physiol. 127(4):1346-1353.
Crossref
|
|
|
|
|
Chabane K, Valkoun J (1997). Standardization of RAPD marker technique to determine the diversity of diploid wheat Triticumurartu. 3rd international Triticeae Symposium held in Aleppo-Syria, 4-8 May, 1997.
|
|
|
|
|
Cheng Z, Sameshima K, Chen JK (2002). Genetic variability portent and relationship of kenaf germplasm based on RAPD analysis (in Chinese). Plant Fibers Prod. 1:1-12.
|
|
|
|
|
Cushman JC, De Rocher EJ, Rohnert HJ (1990) Gene expression during adaptation to salt stress. In: Katterman (ed) Environmental Injury to plants. Academic Press. New York. pp. 173-203.
Crossref
|
|
|
|
|
Daudu OAY, Falusi OA, Dangana MC, Abubakar A, Yahya SA, Abejide DR (2015). Collection and Evaluation of Roselle (Hibiscus sabdariffa L.) Germplasm in Nigeria. Afr. J. Food Sci. 9(3):92-96.
Crossref
|
|
|
|
|
Daudu OAY, Falusi OA, Gana SA, Abubakar A, Oluwajobi AO, Dangana MC, Yahya SA (2016). Assessment of Genetic Diversity among Newly Selected Roselle (Hibiscus sabdariffa Linn.) Genotypes in Nigeria Using Rapd-Pcr Molecular Analysis. World J. Agric. Res. 4(3):64-69.
|
|
|
|
|
Degani D, Rowland LJ, Saunders JA, Hokanson SC, Ogden EL Goldhirsh AG (2001). A comparison of genetic relationship measures in strawberry based on AFLPs, RAPDs, and pedigree data. Euphytica 117:1-12.
Crossref
|
|
|
|
|
Khaje Hosseini M, Powell AA, Bingham IJ (2003). The interaction between salinity stress and seed Vigor during germination of soybean seed. Seed Sci. Technol. 1:715-725.
Crossref
|
|
|
|
|
Larcher W (2001) Physiological Plant Ecology. Springer Verlag Berlin Heidelberg New York. Germany P 505
|
|
|
|
|
Levi A, Rowland LJ (1997). Identifying blue berry cultivars and evaluating their genetic relationship using randomly amplified polymorphic DNA (RAPD) and simple sequence repeat - (SSR-) anchored primers. J. Am. Soc. Hort. Sci. 122(l):74-78.
|
|
|
|
|
Maxwell SE, Delaney HD (1989). Designing Experiments and Analyzing Data, Wadsworth, Belmont, CA. pp. 241-260.
|
|
|
|
|
Nei M (1978). Estimation of average heteroz-ygosity and genetic distance from a small number of individuals. Genetics 89:583-590.
|
|
|
|
|
Netto AT, Campostrini E, de Oliveira JG, Bressan-Smith RE (2005). Photosynthetic pigments, nitrogen, chlorophyll a fluorescence and SPAD-502 readings in coffee leaves. Sci. Horticult. 104(2):199-209.
Crossref
|
|
|
|
|
Neuman PM, Van Volkenburgh E, Clelamd RE (1988). Salinity stress inhibits bean leaf expansion by reducing turgor, not wall extensibility. Plant Physiol. 88:233-237.
Crossref
|
|
|
|
|
Omalsaad AKM, Aminu I, Murshida AJ, Zahira Y, Mohamad O (2014). Genetic relationship between roselle (Hibiscus sabdariffa L.) and kenaf (Hibiscus cannabinus L.) accessions through optimization of PCR based RAPD method. Emirate J. Food Agric. 26(3):247-258.
|
|
|
|
|
Prasad MP (2014). Molecular characterization and genetic diversity determination of Hibiscus species using RAPD molecular markers. Asian J. Plant Sci. Res. 4(3):50-56.
|
|
|
|
|
Passioura JB (1996). Drought and drought tolerance. Plant Growth Regulation 20(2):79-83.
Crossref
|
|
|
|
|
Passioura JB (2007). The drought environment: physical, biological and agricultural perspectives. J. Exp. Bot. 58(2):113-117.
Crossref
|
|
|
|
|
Premachandra GS, Hahn DT, Rhodes D, Joly RJ (1995). Leaf water relations and solute accumulation in two grain sorghum lines exhibiting contrasting drought tolerance. J. Exp. Botany 46(12):1833-1841.
Crossref
|
|
|
|
|
Ragot M, Hoisington DA (1993). Molecular markers for plant breeding: Comparisons of RFLP and RAPD genotyping costs. Theo. Appl. Genet. 86:965-984.
Crossref
|
|
|
|
|
Ramanjulu S, Bartels D (2002). Drought and desiccation induced modulation of gene expression in plants. Plant Cell Environ. 25(2):141-151.
Crossref
|
|
|
|
|
Rogers SO, Bendich AJ (1985). Extraction of DNA from milligram amounts of fresh herbarium and mummified plant tissues. Plant Mol. Biol. 5:69-76.
Crossref
|
|
|
|
|
Seeruttum B, Sanmukiya VMR (2013). Molecular characterisation of some Hibiscus species cultivated in Mauritius. Int. J. Life Sci. Biotechnol. Pharma Res. 2(3):358-366.
|
|
|
|
|
Welsh J, McCeleand M (1990). Fingerprinting genomes using PCR with arbitrary primers. Nucleic Acids Res. 18:7213-7218.
Crossref
|
|
|
|
|
Weising K, Nybom H, Wolff K, Meyer W (1995). DNA Fingerprinting in plant and Fungi. CRC Press, Inc.
|
|
|
|
|
Williams JGK, Kubelik AR, Livak KJ, Rafalski A, Tingey SV (1990). DNA polymorphisms amplified by arbitrary primers are useful as genetic markers Nucleic Acids Res. 18:6531-6535.
Crossref
|
|
|
|
|
Yang X, Quiros C (1993). Identification and classification of celery cultivars with RAPD markers. Theor. Appl. Genet. 86:205-212.
Crossref
|
|
|
|
|
Zehdi S, Sakka H, Rhouma AOM, Salem A, Marrakchi M, Trifi M (2004). Analysis of Tunisian date palm germplasm using simple sequence repeatprimers. Afr. J. Biotechnol. 3:215-219.
Crossref
|
|
