ABSTRACT
The type of gene action conferring combined resistance to maize streak virus (MSV) and maize stripe virus (MStV) was investigated in a Quality Protein Maize (QPM) population, ART/98/SW6-OB. Full-sib and half-sib progenies generated using North Carolina Design I mating scheme were evaluated in 2015 and 2016 in two locations endemic to the viral diseases in Nigeria. Serological study was also conducted on the progenies to validate the disease scores on the field. Significant mean square of genotypes was observed for MSV severity, MStV incidence and severity, plant height and days to flowering, while mean square of environment by females in males’ interaction was significant for grain yield and MSV incidence. Estimates of additive variances were larger than dominance variance for grain yield and MSV severity with moderate to high narrow-sense heritability estimates. This indicates that there is adequate genetic variability for improving grain yield and resistance to MSV in the maize population. The mean titre values of MSV and MStV for the progenies were significantly higher than the healthy control. MSV and MStV incidence were negatively correlated to days to 50% silking. Titre values were negatively correlated with grain yield but positively correlated with MSV and MStV incidence. Recurrent selection method that capitalizes on both additive and dominance variances would be effective in improving the population for grain yield and resistance to MSV and MStV diseases.
Key words: Correlation, gene action, maize streak virus, maize stripe virus, resistance, North Carolina design.
The development of quality protein maize (QPM) which contains twice the levels of tryptophan and lysine in most normal endosperm maize has brought a great hope for human and animal nutrition (Akande and Lamidi, 2006). Despite this good attribute, QPM varieties are threatened by downy mildew and Maize Streak Virus (MSV) diseases among other constraints (Mariote, 2007).
Maize (Zea mays L.) has been reported to be a natural host for more than 30 viruses (Lapierre and Signoret, 2004). Maize stripe virus (MStV) and MSV are two of the biotic factors responsible for reduced yield in maize in Africa (Shepherd et al., 2010). MSV, a geminivirus, indigenous of Africa, is transmitted by at least eleven Cicadulina species with C. mbila as the main vector. The symptoms are characterized by broken to almost continuous chlorotic stripes centered on tertiary leaf veins (Pinner et al., 1988).
Maize plants infected within the first three weeks after emergence become severely stunted, producing abnormal cobs or giving no yield at all (Pinner et al., 1988). MStV is a tenuivirus group transmitted by the maize delphacid, Peregrinus maidis (Ashmead) (Homoptera: Delphacidae) in a persistent-propagative manner (Tsai and Brown, 1994). MStV is also transmitted through the egg of the planthopper vector (Tsai and Zitter, 1982). Initial symptoms of MStV are fine chlorotic stripings between leaf veins which later develop into continuous chlorotic stripes of varying width and intensity (Tsai, 1975). Young plants infected at the early growth stage often are stunted with twisted whorl leaves (Tsai, 1975). Both MSV and MStV have been reported to cause up to 30 to 100% reduction in maize yield (Alegbejo et al., 2002; Roca De Doyle et al., 2007).
Although integrated pest management (1PM) has been recommended as a viable option of control, the most appropriate, environmentally safe and economically viable method to minimize damage caused by maize viruses is host plant resistance (Thottappilly et al., 1993; Alegbejo et al., 2002; Lagat et al., 2008).
In 1975, researchers at the International Institute of Tropical Agriculture (IITA), Nigeria detected resistance to MSV in cv. Tropical Zea Yellow (TZY), after it was then improved through mass selection and transferred to the most productive varieties (Soto et al., 1982). Although Centro Internacional de Mejoramiento de Maíz y Trigo (CIMMYT) and IITA have worked extensively on breeding for resistance to MSV, global climate change and favorable temperatures for the planthopper vector (Laodelphax steriatellus) may have exacerbated the problem.
Disease diagnosis based on symptoms on the plants on the field is unreliable because different viruses may cause similar symptoms and different symptoms may be induced by one virus. Therefore, use of Enzyme-linked Immunosorbent Assay (ELISA), a serological-based technique is important in validating the observations recorded on the field (Kumar, 2009; Sharma and Misra, 2011).
Knowledge of the type of gene action conferring resistance is important in improving crop populations for resistance to the diseases. This enables the breeder to know the type of selection method to adopt and determine the extent of improvement attainable in a breeding program. Estimates of heritability and genetic advance help to predict response from selection (Holland et al., 2003).
Conflicting results have so far been reported on gene actions conferring resistance to MSV. Engelbrecht (1975) found that five dominant genes were involved in resistance to MSV, but Kim et al. (1989) reported that resistance in inbred IB32 was quantitatively inherited through additive gene action of two or three major genes and some modifiers.
DeVries (ISAAA, 1999) pointed out that MSV is known to be controlled by one major gene with several modifiers. Asea (2005) reported mostly dominance gene action for MSV. Lorroki (2009) reported that MSV resistance is controlled by additive gene effects with dominance x dominance epistatic interaction. Dintinger et al. (2005) reported that resistance to MStV is quantitative. This study therefore aims at investigating the type of gene action conferring combined resistance to both MSV and MStV in a QPM population with a view of improving it for dual resistance to both viral diseases.
Generation of progenies
The maize population used in this study was a QPM population, ART/98/SW6-OB. It is a flint-dent, intermediate maturing, and white-grained maize population adapted to the forest zone. 200 non-inbred (S0) plants were randomly selected from the maize population and used to produce both full- and half-sib progenies using the North Carolina Design I (NCD I) mating scheme of Comstock and Robinson (1952). Forty individual S0 plants designated as male parents were each crossed to four different individuals designated as female to produce one hundred and sixty progenies for evaluation. To enhance flower synchronization, male parents were planted 3 days after female parents. To reduce replication size and increase precision of the experiment, the individuals were grouped into sets by males. Five sets of eight males each or 32 full-sib progenies were considered for the experiment.
Field evaluation
The 160 progenies were evaluated under natural infection by MSV and MStV in 2015 and 2016 at Ilora (Lat. 7o 81'N, Long.3o 82'E) in the transition zone between rain forest and derived savanna and Ikenne (Lat. 6o 54'N, Long.03o 42'E) in the humid forest both in Southwestern zone of Nigeria, a zone well known to be endemic to various maize diseases in Nigeria. The evaluations in both years were carried out towards the late season (July) when viral infections reach the peak (Fajemisin, 2003). A randomized incomplete block design with three replications was used for evaluation in each location. A single row plot of 3m length and plant spacing of 0.75m between rows and 0.25m within rows was used. Two seeds were sown per hole and later thinned to one plant per hill at three weeks after planting (WAP) to get a maximum of 13 plants per plot and a plant density of 53,333 plants ha-1. Other cultural practices carried out at both location included weeding and fertilizer application. Paraquat and primextra herbicide were sprayed a day after planting at the rate of 3l/ha. One hand weeding was done at four weeks after planting, while supplementary herbicide spray was done at flowering using paraquat alone. A basal application of compound fertilizer (NPK 20:10:10) was applied at 2WAP at a rate of 60kg N/ha and Urea at 6WAP at 60kg N/ha for optimum plant growth (Oloyede-Kamiyo, 2013).
Detection of MSV and MStV using ELISA technique
Symptomatic and asymptomatic leaf samples were collected from the 160 maize progenies before tasseling. Double antibody sandwich (DAS) ELISA was used to detect the presence of MSVand MStV in the progenies generated. The antibodies to the two viruses were obtained from Agdia-Bioford Inc, Elkhart, Indiana, USA. For each virus, microtitre wells of ELISA plates were loaded with 100 μl of capture antibody and the plates incubated for 4 h at room temperature (25oC±1.0). Plates were washed three times with 1X phosphate buffer saline-tween 20 (PBST). Approximately, 1 g of leaf sample was grinded in 1 ml of Agdia’s general extract buffer (GEB). Extracted samples were coated into the duplicate wells of the microtitre plates at 100 μl per well. Absorbance values were read at 405 nm using a Microtitre Plate Reader (Biotek, ELx800). Samples were considered positive when the values of the test sample were twice the mean value of the sap of healthy plant or negative controls.
Data collection
Data collected at both locations included days to 50% silking estimated as days from planting to the day when 50% of the plants in a plot develop silk. Plant and ear height were measured from five competitive plants per plot as distance from ground level to the base of the tassel, and to the node bearing the first ear respectively. Husk cover rating was on scale of 1 to 9, 1 representing tight and long husk cover, while 9 represents short, very loose and opened husk. Ear aspect was also rated on scale of 1 to 9, 1 representing very clean, well filled ears with well aligned kernel rows, while 9 represents diseased and poorly filled ears. Field weight was determined by weighing harvested ears per plot in kilogram. The moisture content of grains at harvest was determined using a maize moisture meter. Grain yield (t/ha) was obtained as field weight adjusted to 14% moisture content.
Grain yield (t/ha) = (FWT (kg)/Plot size (m2)) x [(100 – moisture content) x 10,000 x SP]/ (100-14) x 1000.
SP = Shelling percentage (weight of grain expressed as a percentage of ear weight)
FWT = Field weight
Severity of streak and stripe was scored on a scale of 1 to 5 based on the extent of symptoms observed on leaves, 1 represents plants with very few or no streak or stripe symptoms, while 5 represents plants with very severe streaking or striping (75% of leaf area). The incidence of virus was determined by counting the number of infected plants (by streak or stripe virus) and expressed as a percentage of the total plant stand per plot.
Data analyses
Means and ranges were estimated. Percentage data were transformed using arcsine transformation before analysis (Steel and Torrie, 1960). Years and locations were pooled as environment. The progenies were referred to as genotypes in the analysis. Analysis of variance for North Carolina Design I (NCD I) was performed using PROC GLM of SAS (Version 9.1). Random model was assumed for the analysis (genotypes and environments were random). The model for NCD I for one environment is:
Yijk = u + mi + fij + rk + eijk (Hallauer et al., 2010)
where u is the mean, mi is the effect of the ith male, fij is the effect of the jth female mated to the ith male, rk is the replication effect, and eijk is the experimental error. After analyzing for each season, Bartlett test for homogeneity of variance was carried out to test for significance between the two seasons. Data were then pooled over sets and over environments and analysed. The form of analysis of variance pooled over sets and over environments (seasons and locations) is as shown in Table 1. Mean square (MS) of ‘males within sets’ was tested with MS due to ‘environment x males in sets’ interaction and ‘females in males in sets’. MS of ‘females in male within sets’ and MS due to ‘environment x males in sets’ interaction were tested by MS due to ‘environment x females in males in sets’ interaction. MS of ‘environment x females in males in sets interaction was tested with MS error (Hallauer et al., 2010). Additive variance (σ2a), dominance variance (σ2d) and their interactions with environments were estimated from mean squares of ANOVA (Table 1) as:
σ2m = (M5-M4-M3+M2)/ref, σ2f/m/s = (M4-M2)/re
σ2a = 4σ2m;
σ2d = 4(σ2f/m- σ2m)
σ2m = (M5-M4-M3+M2)/ref,
σ2f/m/s = (M4-M2)/re
σ2e/m/s = M3-M2)/rf;
σ2 e/f/m/s =(M2-M1)/r
Narrow-sense heritability (NSH) and its standard error were calculated according to Hallauer and Miranda (1988). Phenotypic and genotypic correlation coefficients were computed using variance-covariance matrix and estimates of genotypic and phenotypic variances as described by Falconer (1996).
Ranges were high for all the traits studied (Table 2). The wide ranges suggested wide variability for these traits in the maize population. MStV incidence was much higher (37.8%) than MSV incidence (2.9%). MSV and MStV incidence were much higher in Ilora than Ikenne in both 2015 and 2016 (Figure 1 and 2). MSV incidence was lower in both locations in both years compared to MStV incidence (Figure 1) indicating that MSV was not as prevalent in the locations and seasons under study as MStV. Taiwo et al. (2006) also reported low incidence of MSV (0 to 20%) and maize mottle/chlorotic stunt virus in and around Lagos state, Nigeria. This suggests that disease incidence and severity will differ from year to year and from location to location. The moderate grain yield (1.84 ton/ha) despite the high MStV incidence suggested that the maize population has some level of tolerance for MStV infection.
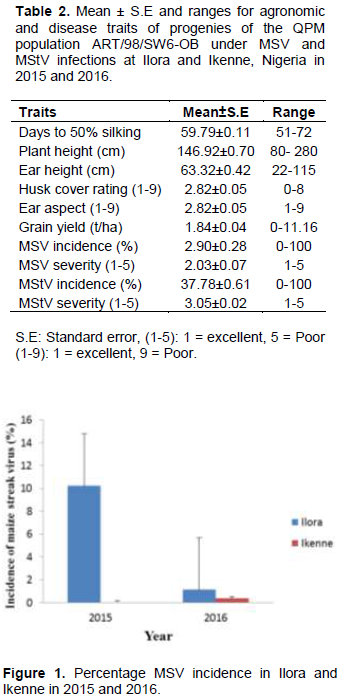
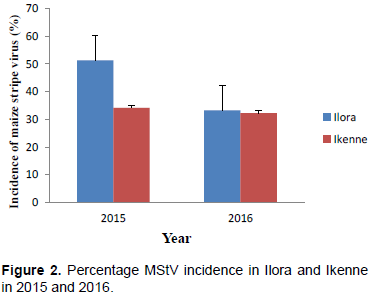
Karavina et al. (2014) also identified some hybrids with multiple resistance to turcicum leaf blight and MSV. QPM varieties are known to be more vulnerable to diseases because of the soft floury endosperm of the Opaque-2 maize which foster fungal growth (NRC, 1988). Bartlett test for homogeniety of variance showed that some traits were significant in the two seasons, while some were not (Table 3).
Combined analysis of variance showed significant mean squares for males in set for days to silking, plant height and MSV severity (Table 4). Mean square (MS) for ‘females within males in set was significant for MStV incidence and severity and most of the agronomic traits except grain yield and ear aspect. MS of ‘environment by female in male in set’ interaction was significant for ear aspect, grain yield, husk cover rating, MSV incidence and MStV incidence and severity. MS of ‘environment by male in set interaction’ was significant for only MStV severity among the disease traits (Table 4).
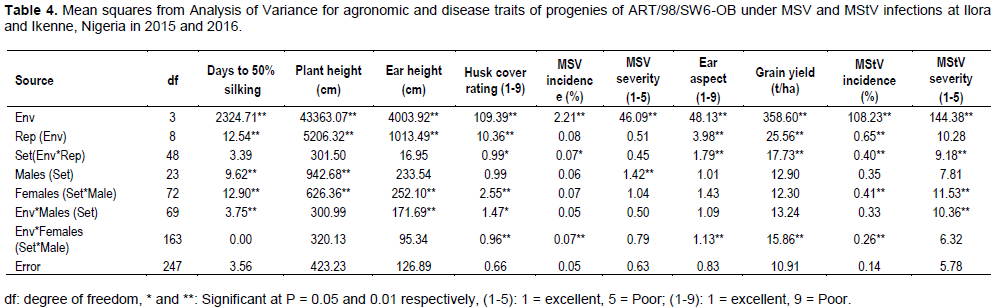
Ige et al. (2017) reported significant MS for GCA by environment and SCA by genotype x environment interactions for maize streak severity. The significant MS of genotypes for plant height, days to 50% silking and the disease traits indicated that the characters are under genetic control and improvement could be made on them. Similar observation was made by Ininda et al. (2006) and Asea (2005) for MSV score.
Dintinger et al. (2005) also reported significant MS of genotypes for maize stripe incidence and severity. Rodier et al. (1995) reported that resistance to MSV in SI and S2 lines derived from population CVR3-C3 was under genetic control with major genes controlling high to complete resistance and minor genes controlling partial resistance. However, Akande and Lamidi (2006) working on three fungal diseases reported on susceptibility of eight QPM varieties to varying degrees with no significant varietal differences. Estimate of dominance variance were larger than additive variance for most of the traits studied except for grain yield and MSV severity (Table 5). This corroborates the finding of Kim et al. (1989) who reported that resistance to MSV in 1B32 inbred line from IITA is controlled quantitatively, mainly by additive gene action, with relatively small (2 to 3) number of genes involved.
Studies conducted by Pixley et al. (1997) showed that additive effects are important for resistance. Lorroki (2009) working on six generations also reported that MSV is controlled by additive gene effect with dominance x dominance epistatic interaction. The number of effective factors was estimated to be between 2 to 7 genes. Ige (2016) reported that additive gene effect was preponderant for MSV severity, while non-additive gene effect was important for grain yield. Sibiya et al. (2013) however reported that additive gene effect was preponderant for grain yield.
Gichuru (2013) stated that varying gene actions controlling grain yield is dependent on the parent and the environment under consideration. Additive genetic variance for husk cover rating and ear aspect were negative and were therefore equated to zero. Dominance variance being larger than additive variance for MSV incidence, MStV incidence and severity with average degree of dominance being above unity has also been reported (Dintinger et al., 2005; Rodier et al.,1995; Asea, 2005).
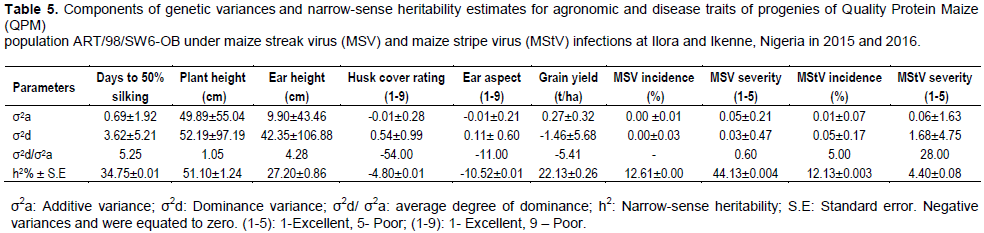
Dintinger et al. (2005) reported that resistance to MStV is quantitative and may vary in relation to cumulative number of innoculative planthoppers by hour. Rodier et al. (1995) also reported that dominance was important for resistance to MStV. Asea (2005) reported mostly dominance gene action for MSV. Out of the 10 traits studied, average degree of dominance (σ2d/ σ2a) was above unity for five of the traits. No dominance was observed for husk cover rating, ear aspect, grain yield and MSV incidence, while partial dominance was observed for MSV severity (Table 5).
The report of Mariote (2007) corroborates the finding in the present study that resistance to MSV is controlled by partial dominance. This result is however surprising because this population has not been selected. According to Hallauer and Miranda (1988), in an unselected maize material, the variance for GCA, an indicator of additive genetic variance was larger than the variance for SCA, related to dominance variance. Average degree of dominance being above unity for MStV incidence and severity and the agronomic traits suggests presence of complete dominance for the genes affecting these traits or possible occurrence of over-dominance at some loci (Hallauer and Miranda, 1988).
The strikingly high degree of dominance and very low heritability for MStV severity suggests presence of epistatic variance and considerable portion of genotype by environment interaction variance. This is revealed in the mean square table with environment by male in set interaction being significant for MStV severity. The build-up of dominance variance component may be due to the fact that when there is overdominance, heterozygote is favoured, and both favourable and unfavourable alleles will be accumulated in the population instead of eliminating the unfavourable alleles. The presence of both additive and dominance variance in the maize population, ART/98/SW6-OB, suggests that breeding scheme that capitalizes on both types of gene action such as S1 selection, half-sib or test cross could be used to improve the population for resistance to both maize MSV and MStV diseases.
Narrow-sense heritability (NSH) estimate was moderate for most of the traits. It was least in MStV severity (4.4%) and highest in plant height (51.0%) (Table 5). Among the disease traits, MSV severity had the highest NSH of 44.1%. Ige (2016) also reported high NSH (55.3%) for maize MSV severity. Negative NSH were observed for husk cover rating and ear aspect. The moderate to high heritability, and wide ranges observed in the maize population for grain yield, MSV severity, days to silking and plant height, suggests that there is adequate genetic variation in the population for grain yield and resistance to the MSV and that progress would be made from selection. The heritability value obtained for MSV in this study falls within the range obtained by Asea (2005) on MSV. Soto et al. (1982) also found resistance to be simply inherited and fixable rapidly through breeding for MSV.
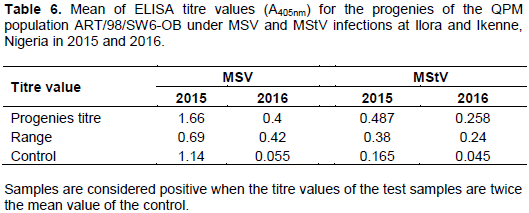
Means of ELISA titre values for MSV and MStV are shown in Table 6. Means of the progenies were significantly higher than the control except for MSV titre in 2015. Ranges of the titre values were moderately wide for both MSV and MStV in each season (Table 6). The mean ELISA titre values being significantly higher than the healthy control revealed the presence of the viruses in the maize population. Some of the progenies tested positive while some tested negative compared with the healthy control (data not shown). The ranges of the titres presented here further indicated that there is variability for resistance to the double viruses in the maize population. ELISA technique has been widely used to detect the presence of viruses in maize with or without disease symptoms (Kumar et al., 2004; Fajinmi et al., 2012).
Estimates of correlation coefficients among traits under MSV and MStV infections are shown in Table 7. Days to 50% silking had negative correlations with plant height (rp= -0.24*) and grain yield (rp = -0.21*, rg = -0.21). Grain yield had positive and significant correlations with plant height (rp = 0.32*, rg = 0.35) but negative correlation with husk cover rating (rp = -0.28*, rg = -0.35). This indicates that tall plants with good husk cover rating tend to have better grain yield. For the relationship among agronomic and disease traits, days to 50% silking had negative genotypic correlations with maize MSV incidence (-0.36) and MStV incidence (-0.62).

Plant height also had negative correlations with disease incidence except with MStV incidence (rg =0.63) although phenotypic correlation was not significant for MSV and MStV incidence. The significant negative phenotypic correlation between plant height and disease incidence was also observed by Mariote (2007) and Asea (2005). Mariote (2007) reported negative phenotypic correlation between MSV and plant height, while Asea (2005) reported negative correlation between MSV and days to silking.
Grain yield was significantly correlated with MSV titre (rp = -0.32*, rg = -0.34) and MStV titre (rp = -0.33**, rg = -0.49). The negative correlation between grain yield and the titre values indicated that the lower the titre values the higher the yield. The positive phenotypic correlation between grainyield and MSV incidence and severity was also reported by Mariote (2007). Lorroki (2009) reported that MSV disease significantly reduced stover dry matter and grain yield with yield loss ranging between 8 to 48%. Strong relationship existed between MSV incidence and severity (rp =0.69**, rg = 0.80).
MSV and MStV incidence also had strong correlation with MSV and MStV titres. The strong positive relationship between the titre values and MSV and MStV incidence further supports the ability of ELISA technique to detect various viral infections. The significant correlation between MSV incidence and severity indicates that the higher the incidence, the higher the severity. Similar observation was made between MStV incidence and severity (Dintinger et al., 2005). Highly significant correlation also existed between the two titres (rp =0.89**, rg = 0.84). This is an indication of possible interaction between the two viruses and among diseases generally on plants. Karavina et al. (2014) reported that yield of MSV-infected plants was not significantly different from yield of turcicum leaf blight-infected plants.
The larger additive variance estimates and moderate to high heritability for grain yield and MSV severity with wide ranges indicates that improving the maize population for grain yield and MSV resistance would be successful. However, progress in improving the population for MStV resistance would be slow due to the low heritability estimates for MStV scores. Breeding scheme that capitalizes on both additive and dominance gene action such as S1 selection, half-sib or test cross could be used to improve the population for resistance to both maize MSV and MStV diseases.
The authors have not declared any conflict of interests.
REFERENCES
|
Alegbejo MD, Olojede SO, Kashina BD, Abo ME (2002). Maize streak virus in Africa: Distribution, transmission, epidemiology, economic significance and management strategies. Journal of Sustainable Agriculture, 19(4):35-45.
Crossref
|
|
|
|
Akande SR, Lamidi GO (2006). Performance of quality protein maize varieties and disease reaction in the derived-savanna agro-ecology of South-West Nigeria. African Journal of Biotechnology, 5(19):1744 -1748.
|
|
|
|
|
Asea G (2005). Genetic characterization of partial resistance and comparative strategies for improvement of host-resistance to multiple foliar pathogens of maize. Ph.D Thesis. Horticulture and Crop Science The Ohio State University xvii+171p.
|
|
|
|
|
Comstock RE, Robinson HF (1952). Estimation of average dominance of genes. In. J.W. Gowen, (Ed) Heterosis. Iowa State University press, Ames, Iowa. pp. 494-516.
|
|
|
|
|
Dintinger J, Boissot N, Chiroleu F, Hamon P, Reynaud B (2005). Evaluation of maize inbreds for maize stripe virus and maize mosaic virus resistance: Disease progress in relation to time and the cumulative number of planthoppers. Phytopathology, 95:600-607.
Crossref
|
|
|
|
|
Engelbrecht GC (1975). Streak, a major threat? South Africa Department of Agricultural Technical Communication, 132:101-103.
|
|
|
|
|
Falconer DS (1996). Introduction to quantitative genetics. 4th edition. New York: Benjamin Cummings, Longman Inc.
|
|
|
|
|
Fajemisin JM (2003). Overview of maize viruses in sub-Saharan Africa.In: JH Hughes and J. Odu (eds). Plant virology in sub-Saharan Africa. International Institute of Tropiucal Agriculture Conference, Ilesa, Nigeria, pp. 158-171.
|
|
|
|
|
Fajinmi A, Dokunmu AO, Akheituamen DO, Onanuga KA (2012). Incidence and infection rate of maise streak viru disease by Cicadulina triangular on maize plants and its distribution from the lowest diseased leaf under tropical conditions. Archives of Phytopathology Protection, 45(13):1-8.
|
|
|
|
|
Gichuru LJ (2013). Breeding investigations on utility of maize streak virus resistant germplasm for hybrid development in the tropics Ph.D Thesis. School of Agricultural, Earth and Environmental Sciences College of Agriculture, Engineering and Science .University of KwaZulu-Natal, Pietermaritzburg, Republic of South Africa. xvii+226pp.
|
|
|
|
|
Hallauer AR, Miranda JB (1988). Quantitative genetics in maize breeding. Iowa State University Press. Ames, Iowa.
|
|
|
|
|
Hallauer AR, Carena MJ Miranda JB (2010). Quantitative genetics in maize breeding. Springer New York. Dordrecht Heidelberg London, 680 p.
|
|
|
|
|
Holland JB, Nyquist WE, Cervantes-Martinez CT (2003). Estimating and interpreting heritability for plant breeding: an update. Plant Breeding Reviews, 22:9-13.
|
|
|
|
|
Ige AD (2016). Combining ability, heterosis and heritability of agronomic traits and resistance to maize streak virus in maize inbred lines. M. Phil. Thesis. Faculty of Agriculture, Department of Crop and Soil Sciences. Kwame Nkrumah University of Science and Technology, Kumasi, Ghana. xix + 92p.
|
|
|
|
|
Ige AD, Akromah R, Oppong A and Adejumobi II (2017). Combining ability of tropical early maize (Zea mays L.) inbred lines for grain yield and resistance to maize streak virus disease. Journal of Plant Breeding and Crop Science, 9(10):186-194.
Crossref
|
|
|
|
|
Ininda J, Gichuru L, Njuguna JGM, Lorroki P (2006). Performance of three-way cross hybrids for agronomic traits and resistance to maize streak virus diseases in Kenya. Africa Crop Science Journal, 14(4):287-296.
|
|
|
|
|
International Service for the Acquisition of Agri-biotech Applications (ISAAA) (1999). Briefs No. 16. Advances in maize streak virus research in Eastern and Southern Africa, workshop Report, 15-17 September KARI and ISAAA africenter.
|
|
|
|
|
Karavina C, Mandumbu R, Mukaro R (2014). Evaluation of three-way maize (Zea mays L.) hybrids for yield and resistance to maize streak virus and turcicum leaf blight diseases. Journal of Animal Plant Science, 24(1):216-220.
|
|
|
|
|
Kim SK, Effron Y, Fajemisin JM, Buddenhagen IW (1989). Mode of gene action for resistance in maize to maize streak virus. Crop Science, 29:890-894.
Crossref
|
|
|
|
|
Kumar PL, Jones AT, Waliyar F (Eds) (2004). Serological and nucleic acid-based methods for the detection of plant viruses. Virology and Mycotoxin Diagnostics Laboratory. International Crop Research Institute for the Semi-Arid Tropics. Patancheru 502324, India. P 109.
|
|
|
|
|
Kumar PL (2009). ELISA and RT-PCR Methods for the detection of plant viruses. IITA, Ibadan. pp. 55-106.
|
|
|
|
|
Lagat M, Darson J, Kimani M, Kuria A (2008). Quantitative trait loci for resistance to maize streak virus disease in maize genotypes used in hybrid development. African Journal of Biotechnology, 7(14):2573-2577.
|
|
|
|
|
Lapierre H, Signoret PA (2004). Viruses and virus diseases of Poaceae (Gramineae). INRA. Paris, France.
|
|
|
|
|
Lorroki P (2009). Genetic analysis of resistance to maize streak virus. Ph.D Thesis. Department of Plant Science and Crop Protection. College of Agriculture and Veterinary Science. University of Nairobi. Xii + 105pp.
|
|
|
|
|
Mariote D (2007). Response to Selection for downy mildew (Peronosclerospora sorghi) and maize streak virus resistance in three Quality Protein Maize populations in Mozambique. Ph. D Thesis.
|
|
|
|
|
National Research Council (1988). Quality Protein Maize. National Academy Press. Washington D.C. USA. pp. 41-54.
|
|
|
|
|
Pinner MS, Markham PG, Markham RH, Dekker I (1988). Characterization of maize streak virus: description of strains; symptom. Plant Pathology, 37(1):74 -87.
Crossref
|
|
|
|
|
Pixley KV, Jewell DC, Schechert A, Welz G (1997). Recent maize streak disease research at C1MMYT Zimbabwe. Proceeding of the 1st International Maize streak disease symposium. Hazyview, Mpumalangu September 9-11, 1997.
|
|
|
|
|
Roca de Doyle MM, Autrey LJC, Jones P (2007). Purification, characterization and serological properties of two virus isolates associated with the maize stripe disease in Mauritius. Plant Pathology, 41(3):325-334.
Crossref
|
|
|
|
|
Rodier A, Marchand JL, Herve Y (1995). Breeding maize lines for complete and partial resistance to maize streak virus (MSV). Euphytica, 81(1):57- 70.
Crossref
|
|
|
|
|
Sharma K, Misra RS (2011). Molecular approaches towards analyzing the viruses infecting maize (Zea mays L.). Journal of General and Molecular Virology, 3(1):1-17.
|
|
|
|
|
Shepherd DN, Martin DP, Walt EVD, Dent K, Varsani A, Rybicki EP (2010). Maize streak virus: an old and complex 'emerging' pathogen. Molecular Plant Pathology, 1:11-12.
Crossref
|
|
|
|
|
Sibiya J, Tongoona P, Derera J (2013). Combining ability and GGE biplot analyses for resistance to Northern leaf blight in tropical and sub-tropical elite maize inbred lines. Euphytica, 191:245-257.
Crossref
|
|
|
|
|
Soto PE, Buddenhagen IW, Asnani VL (1982). Development of streak virus-resistant maize population through improved challenge and selection methods. Annals of Applied Biology, 100:539-546.
Crossref
|
|
|
|
|
Steel RGD, Torrie JH (1960). Principles and procedures of statistics. McGraw- Hill Book Co. New York, Toronto. London 481p.
|
|
|
|
|
Taiwo MA, Hughes Jd'A, Oke KE (2006). Studies on maize streak virus and maize mottle/chlorotic stunt virus in Lagos, Nigeria. Plant Disease 90(2):199-202.
Crossref
|
|
|
|
|
Thottappilly G, Bosque-Pérez NA, Rossel HW (1993). Viruses and virus diseases of maize in tropical Africa. Plant Pathology, 42:494-509.
Crossref
|
|
|
|
|
Tsai JH (1975). Occurrence of a corn disease in Florida transmitted by Peregrinus maidis. Plant Disease Reporter, 59:830-833.
|
|
|
|
|
Tsai JH, Brown LG (1994). Maize Stripe Tenuivirus. Plant Pathology Circular No. 363.
|
|
|
|
|
Tsai JH, Zitter TA (1982) Transmission characteristics of maize stripe virus by the corn delphacid. Journal of Economics Entomology, 75:397-341.
Crossref
|
|