ABSTRACT
In recent years very virulent (VV) IBDV strains and classical (CV) IBDV strains re-emerged and caused devastating outbreaks in different parts of the world. In this study, genetic evolution of fifteen IBDVs collected in Senegal in 1979, 1999, 2007, 2012, 2013 and 2014 was characterized to gain information for a better control of IBD. Following RT-PCR, nucleotide sequence of the VP2 hypervariable region was determined and compared with sequences available in GenBank. Phylogenetic analysis showed that the viruses diverged into two genotypes: Very virulent (VV) IBDV and classical virulent (CV) IBDV. The Senegalese field strains of the first genotype (VV) IBDV had 98.9 to 100% identity among themselves, whereas their identity with reported Nigerian (VV) IBDVs ranged between 96.7 and 99%. The close phylogenetic relationship of the Senegalese and Nigerian strains suggests that they likely derived from a common ancestor. In the phylogenetic tree, all the Senegalese (VV) IBDV strains belonged to the African very virulent types (VV2). The genotyping of senegalese field IBDV strains indicated that the majority of viruses circulating in Senegal are (VV) IBDVs and highlights a genetic stability.
Key words: Infectious bursal disease, phylogenetic analysis, Senegal.
Infectious bursal disease (IBD) is a highly contagious acute viral disease of young chickens (3 to 6 weeks old) that causes immunosuppression by damaging the bursa of Fabricius. The disease is either fatal or causes impaired growth of young chickens, resulting in significant economic losses for the poultry industry (Islam et al., 2005). Infectious bursal disease virus (IBDV) is a member of the genus Avibirnavirus of the family Birnaviridae. There are two distinct serotypes of IBDV, 1 and 2. Serotype 1 viruses are pathogenic to chickens, while serotype 2 viruses are non-pathogenic to chickens (Ismail et al.,1998). IBDV can also be grouped into pathotypes based on their pathogenicity in chickens: They are referred to as classical virulent (CV), antigenic variant, very virulent (VV) and attenuated (Műller et al., 2003).
IBDV is a double stranded RNA virus with a bisegmented genome. The larger segment A encodes four viral proteins designated as VP2, VP3, VP4 and VP5. The smaller segment B encodes only VP1 which has polymerase activity (Tamura et al., 2011). The hypervariable region (HVR) within VP2, between amino acid residues 206 and 350, has the highest amino acid sequence variation among serotype 1 strains, and the nucleotide and deduced amino acid sequences of this region are widely used for molecular diagnosis and genotyping of IBDVs. This region includes two major sets of hydrophilic amino acids, termed peak A and B, from amino acid positions 210 to 225 and 312 to 324, respectively, with smaller hydrophilic peaks present in-between (Bayliss et al., 1990). So far, the four amino acid changes in VP2 at positions 222A, 256I, 294I, and 299S have been shown to be present in all European-like (VV) IBDVs and serve to differentiated (VV) from (CV) viruses (Zierenberg et al., 2000).
During IBDV replication, the RNA-dependent polymerase (RdRp)-error prone activity potentially leads to a high rate of mutations resulting in antigenic variation in protein domains involved in virus neutralization. This process is thought to be a contributor to the emergence of variant viruses or highly virulent forms seen in the USA and others countries. In the USA, it was demonstrated that the new isolates were affected by antigenic drift against which classical IBDV vaccines were not protective (Snyder et al., 1992), whereas in Europe, the first cases of acute IBDV were described in the 1990s (Dormitorio et al., 1997). Surprisingly, some of these first acute outbreaks occurred in broilers, at the end of the fattening period, at farms where all the necessary hygiene and prophylactic measures had been taken. These findings indicated a dramatic change in the field situation (Van den Berg, 2000).
In Senegal, the first IBD outbreaks were reported in 1975 involving 21 to 60 day old broilers and pullets (Sagna, 1975). Subsequently, IBD has become a major player among infectious diseases of broilers and pullets, despite routine implementation of vaccination. For improved control of this disease in Senegal, genetic evolution of fifteen Senegalese IBDV strains isolated in 1979, 1999, 2007, 2012, 2013 and 2014 was studied. The hypervariable region of the VP2 gene of these field strains was sequenced and compared to each other and to published sequences obtained from GenBank.
Bursa sample collection and preparation of viral suspensions
For virus detection, the bursa samples were aseptically collected from IBD suspected dead chickens. The samples were collected from commercial broilers and pullet flocks in 1979, 1999, 2007 and during the period from 2012 to February 2014 (Table 1).
The samples were subjected for postmortem examination, placed into individual sterile universal bottles and transported under cold chain to the National Veterinary Research Laboratory, Dakar, Senegal. A freeze-dried live vaccine (HIPRAGUMBORO® CH/80, Hipra laboratory, Spain) containing the CH/80 strain of IBDV was taken as positive control and phosphate buffered saline (PBS) as negative control.
In order to prepare viral suspensions, ten milliliter of PBS (pH 7.2) was added to approximately 100 mg of bursal tissue from each sample and homogenized using a pestle and mortar. The homogenates were centrifuged at 12000 rpm, 4°C for 5 min and the supernatants collected for molecular analysis.
IBDV detection by reverse transcription-polymerase chain reaction (RT-PCR)
The viral RNA was extracted using the RNeasy Plus Mini Kit (QIAGEN) from bursal suspensions according to the manufacturer’s protocol and was subjected to RT-PCR.
A 743-bp segment of the highly variable region of VP2 from nucleotides 737 to 1479 was amplified using primers 743-1 (5′- GCCCAGAGTCTACACCAT-3′) and 743-2 (5′-CCCGGATTATGTCTTTGA- 3′) (Jackwood et al., 2011). These primers used for both RT and PCR are located in a conserved sequence region and amplify this portion of genome segment A from all serotype 1 IBDV strains including vvIBDV and non-vvIBDV. They do not amplify genome segment A from serotype 2 viruses. The RT-PCR reactions were conducted using the QIAGEN One Step RT-PCR kit according to the manufacturer's instructions. For one test, the reaction mixture consisted of 15 µl RNase free water, 5 µl purified RNA, 10 μl PCR Buffer Tampon 5X , 2 µl of 10 mM dNTPs mix, 3 µl of each primer (10 µM), 10 µl of Q 5X Solution and 2 μl of one Step RT-PCR Enzyme mix. PCR tubes were placed in the Bio-Rad MJ Mini Personal Thermal Cycler and subjected to 35 amplification cycles.
The RT incubation was at 50°C for 30 min followed by 95°C for 15 min to activate Taq polymerase enzyme and 35 cycles of 95°C for 30 s, 53°C for 1.5 min and 72°C for 1.0 min. At the end of the 35 PCR cycles, a 7.0 min at 72°C extension period was added.
The PCR products were analysed on 2% agarose gels prepared in Tris-Acetate-EDTA (TAE) buffer and stained with ethidium bromide. Eight micro-liter of the PCR product from each of the tubes were mixed with 2 μl 6X Load Dye and electrophoresed along with a DNA ladder 100 pb (Promega) at a constant 100 V for 60 min in 1 X TAE buffer. Amplified product was viewed under UV light for the expected 743 bp. Before sequencing, the amplified products were purified by “QIAquick PCR Purification Kit” (QIAGEN).
Sequencing and phylogenetic analysis
The RT-PCR products for fifteen selected positive samples were sequenced at the Genomics Platform GeT-Purpan, UDEAR UMR 5165 CNRS/UPS, CHU PURPAN, France on a 3130XL Applied Biosystems capillary sequencer. Samples were first purified using the QIAquick PCR Purification Kit (Qiagen). Sanger sequencing was carried out using the same primers as those used for the PCR. Sequences were aligned with reference strains from the GenBank database using Clustal W and a phylogenetic tree of the nucleotides was constructed using MEGA version 5.10 (Tamura et al., 2011) with up to 1000 bootstrapping replicates. The neighbor joining (NJ) method and Pairwise distance were used.
Accession numbers of sequence data. The nucleotides sequence data reported in this article have been submitted to the GenBank sequence database and have been assigned the accession numbers.
Nucleotide sequence analysis
The nucleotide sequence of the hypervariable domain of VP2 from nucleotide positions 737 to 1479 was determined for the fifteen samples in this study (Table 1) and sequences were submitted to GenBank. Thus, the analysis revealed that the Senegalese field IBDV strains had 98.9 to 100% identity among themselves, whereas their identity with the reported Nigerian (VV) IBDV strains (IBDV33/Abuja. NG/2011, IBDV61/Kaduna. NG/2009, IBDV43/Kaduna. NG/2011, IBDV79/Kaduna. NG/2011, IBDV2/Kaduna. NG/2009, IBDV58/Kaduna. NG/2010) ranged between 96.7 and 99%. These Senegalese field isolates were more close to the Nigerian (VV) IBDV isolates IBDV33/Abuja. NG/2011, IBDV61/Kaduna. NG/2009, IBDV43/Kaduna. NG/2011, IBDV79/Kaduna. NG/2011, IBDV2/Kaduna. NG/2009, IBDV58/Kaduna. NG/2010 (identity up to 99%) than the reported Asian and European (VV) IBDV strains OKYM and Cro-Ig/02 (identity up to 96.9%). The Senegalese field isolates IBDV/DK3/SN/2014 and IBDV/KL1/SN/2013 shared 94.6 to 96.9% identity within themselves. The identity of these Senegalese field isolates with the American and European reported (CV) IBDVs (STC, Cu-1, Vaccine Cevac-IBD-L and Faragher/52-70) ranged between 94.6 and 100% identity. All the fifteen Senegalese field isolates studied had a minimum identity of 68.9% with serotype 2 strain IBDVP234 and a maximum identity of 71.0%.
Deduced amino acid sequence analysis
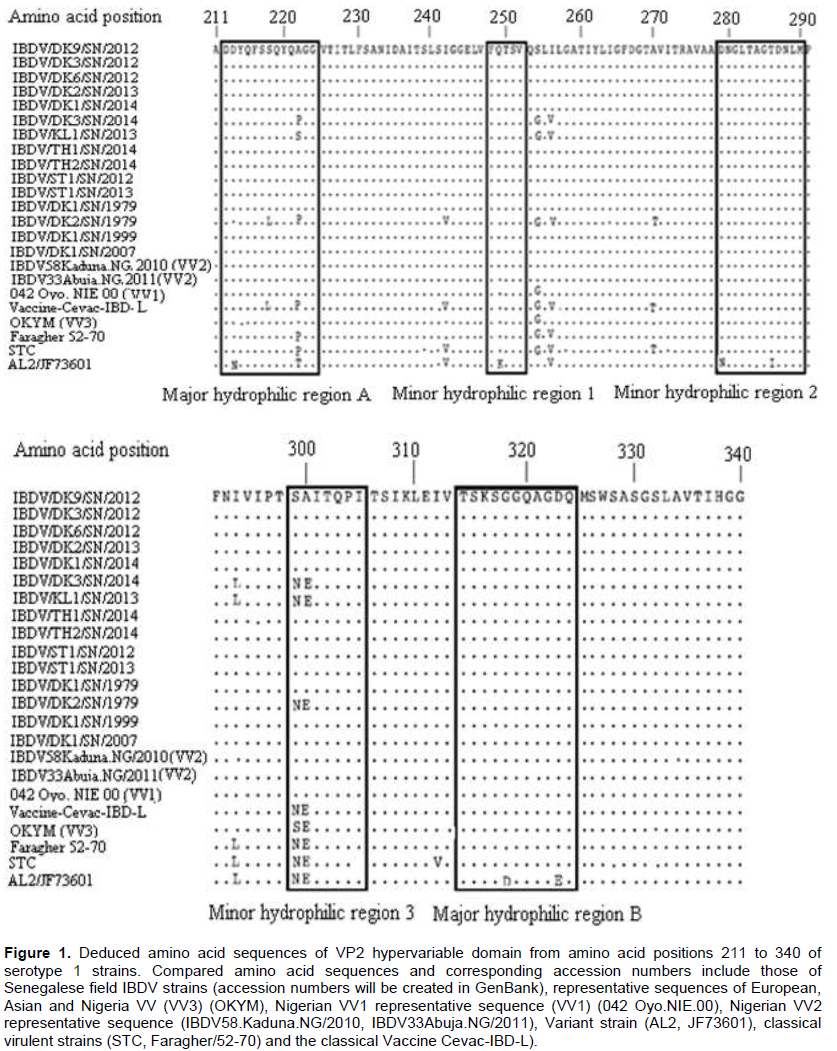
Amino acid sequence analysis revealed 100% sequence similarity among the Senegalese field isolates (Figure 1). Their identity with the Nigerian IBDV strains (IBDV33Abuja. NG/2011, IBDV61/Kaduna. NG/2009, IBDV43Kaduna. NG/2011, IBDV79Kaduna. NG/2011, IBDV2Kaduna. NG/2009, and IBDV58Kaduna. NG/2010) was also 100%. Their similarity with other reported (VV) IBDVs (Cro-Ig/02, OKYM, IBDV80/Kaduna. NG/2011, 050 Oyo. NIE/99, 042 Oyo. NIE/00, 034 Oyo. NIE/99, and 082 Ogun. NIE/97) ranged between 96.9 and 99.2%. The amino acid sequence of the Senegalese field isolates IBDV/DK3/SN/2014 was similar to the strain IBDV/KL1/ SN/2014 (99.2%). The Senegalese field isolate IBDV/DK9/SN/2013, IBDV/DK3/SN/2012, IBDV/DK6/SN/2012, IBDV/DK1/SN/2014, IBDV/ST1/SN/2012, IBDV/TH1/SN/2014, IBDV/ST1/SN/2013, IBDV/DK1/SN/1999, IBDV/DK2/SN/2013, IBDV/DK1/SN/2007, IBDV/DK1/SN/1979 and IBDV/TH2/SN/2014 harbored amino acids 222(A), 242(I), 256(I), 294(I), 299(S) and 300 (A). The strains IBDV/DK3/SN/2014 and IBDV/KL1/SN/2013 had 222(P) or 222(S), 256(V) and 299(N) which allow for the distinction between (VV) and (CV) IBDVs from (CV) IBDVs. All the Senegalese strains had the same amino acid markers at position 253 (Q), 279 (D) and 284(A) (Figure 1).
Phylogenetic analysis
Phylogenetic analysis based on nucleotide sequence revealed that all fifteen Senegalese field isolates fell within the serotype 1 viruses and form two separate genotypes. The (VV) genotype included the Senegalese strains IBDV/DK9/SN/2013, IBDV/DK3/SN/2012, IBDV/DK6/SN/2012, IBDV/DK1/SN/2014, IBDV/ST1/SN/2012, IBDV/TH1/SN/2014, IBDV/ST1/SN/2013, IBDV/DK1/SN/1999, IBDV/DK2/SN/2013, IBDV/DK1/SN/2007, IBDV/DK1/SN/1979 and IBDV/TH2/SN/2014 as well as the reported (VV) IBDVs isolates from Nigeria, Europe and Asia. In the tree, the (VV) genotype was subdivided into three major clusters namely VV1, VV2 and VV3. All these Senegalese (VV) IBDV strains were grouped with Nigerian (VV) IBDV strains (IBDV33/Abuja.NG/2011, IBDV61/Kaduna.NG/2009, IBDV43/Kaduna.NG/2011, IBDV79/Kaduna.NG/2011, IBDV2/Kaduna.NG/2009, and IBDV58/Kaduna.NG/2010) in the cluster VV2. The reported strains 050 Oyo.NIE/99, 042 Oyo.NIE/00, 034 Oyo.NIE/99 and 082 Ogun.NIE/97 formed the cluster VV1 and the reported strains Cro-Ig/02, OKYM, IBDV80/Kaduna.NG/2011 were regrouped in the cluster VV3. In the classical genotype cluster, the Senegalese strains IBDV/DK3/SN/2014 and IBDV/KL1/SN/2013 were closely related to the classical strains STC, Cu-1, Vaccine Cevac-IBD-L and Faragher/52-70 isolated in the U.S.A. and Europe. The (CV) IBDV strain IBDV/DK2/SN/1979 showed 100% nucleotide sequence identity compared with the Cevac-IBD-L vaccine (Figure 2).
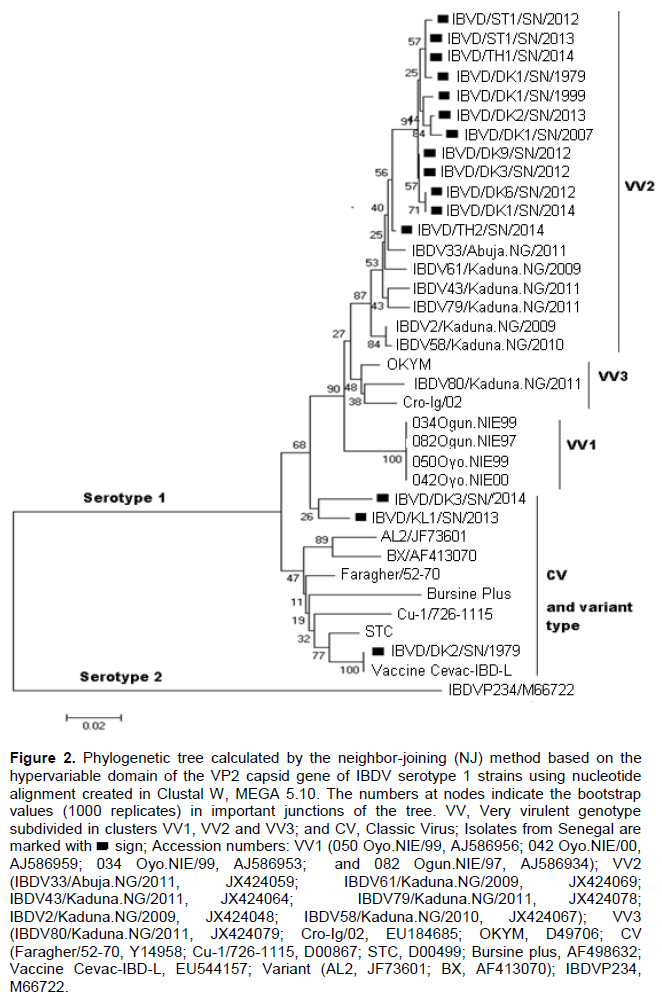
Infectious Bursal disease is one of the major problems faced by modern poultry farming in Senegal and is responsible for considerable economic losses particularly in broiler farms. The first cases of IBD diagnosed clinically were recorded in 1975 in Dakar with an important mortality rate (Sagna, 1975). Since then IBD outbreaks have been noted in broilers and layers chickens in modern poultry farming throughout Senegal. The Senegalese VV strains showed intra-group nucleotide sequence identities within themselves of 98.9 to 100% and amino acid sequence identities of 100%. They were all grouped in the same cluster VV2 with six Nigerian (VV) IBDVs (IBDV33/Abuja.NG/2011, IBDV61/Kaduna.NG/2009, IBDV43/Kaduna.NG/2011, IBDV79/Kaduna.NG/2011, IBDV2/Kaduna.NG/2009 and IBDV58Kaduna.NG/2010). These Senegalese strains were closely related to the reference (VV) IBDV strains from Europe (Cro-Ig/02) and from Asia (OKYM) but belonged to a different lineage. The Senegalese strain IBDV/TH2/SN/2014 and the Nigerian strains IBDV33/Abuja.NG/2011 and IBDV61/Kaduna.NG/2009 were found to share 99 and 100% identity at the nucleotide and amino acid levels, respectively. It could be speculated that the same IBDV strains were exchanged between the two countries or that the outbreaks in the two countries might have a common source. However, the epidemiological links between the outbreaks in the two countries are not known. These findings demonstrate the presence of African (VV) IBDV type in Senegal since 1979. Kasanga et al. (2007) reported that the African genetic lineage may have spread worldwide later on. This suggests that similar viruses clustering in VV2 may exist in other countries in West Africa.
Genetic stability over time within the VP2 hypervariable domain has been detected in Italy (Martin et al., 2007). This is in contrast to the sequence heterogeneity detected among (VV) IBDV isolates from other African countries. Indeed, Adamu (2013) reported in Nigeria a high level of genetic heterogeneity and two distinct genetic clusters, specifically VV1 and VV2. The high degree of sequence diversity among Nigerian (VV) IBDV isolates suggests that West-Africa may be the origin of the newly emerged (VV) IBDV variant found across the Old World (Owoade et al., 2004). In Tanzania, Kasanga et al. (2007) reported the circulation of the (VV) IBDV isolates from both Asian/European lineages.
The phylogenetic analysis showed that the Senegalese strains IBDV/DK3/SN/2014, and IBDV/KL1/SN/2013 belonged to the (CV) genotype. These strains were related to the reference (CV) IBDV from the U.S.A. (STC, Cu-1) and from Europe (Faragher/52-70) but they also clustered in a different sub-lineage. According Sagna (1975) the Cevac-IBD-L vaccine containing intermediate plus strains was widely imported and used by the Senegalese poultry farmers since 1975. And this vaccine preserved an important residual pathogenic power which caused IBD.
Antigenic variation of IBDVs is largely due to mutations occurring in two main hydrophilic regions of VP2, located between amino acid residues 212 to 223 and amino acid residues 314 to 324 (Vakharia et al., 1994). It has been reported that the amino acid composition at position 222 can be used to define groups of IBDVs. Classical viruses typically harbor a proline at position 222, variant viruses a threonine and a serine at position 222 can be found in both classical and variant viruses (Dormitorio et al., 1997). The results revealed that all the Senegalese (VV) IBDV strains contained the putative virulence marker amino acids at positions 222(A), 242(I), 256(I), 294(I) and 299(S) (Figure 2), which have been identified in most (VV) IBDVs (Kasanga et al., 2007). The amino acids at positions 222 (A), 256 (I) and 294 (I) were reported to be unique to all known (VV) IBDV strains (Banda et al., 2004). In addition, these strains retained the amino acid substitution at position 300 (E→A) in the minor hydrophilic region of the VP2 variable domain, as was also observed in the African (VV) IBDVs from Tanzania (Owoade et al., 2004), Zambia (Kasanga et al., 2013) and Nigeria (Adamu et al., 2013). All the Senegalese IBDV strains had the same amino acid sequences at positions 253 (Q), 279 (D) and 284 (A). Similarly, Mawgod et al. (2014) reported the same amino acid sequences at these positions of the VP2 in field strains in Egypt. The amino acids at positions 253 and 284 were found to be responsible for pathogenicity and are unique to highly virulent IBDVs (Brandt et al., 2001).
These observations confirm the circulating of (VV) IBDVs in Senegal with high mortality. Consequently, a variety of freeze-dried live vaccines containing inter-mediate plus IBDV strains are widely used in Senegalese commercial chickens to get better protection against currently circulating (VV) IBDV. Thus, the pathogenicity and protective efficacy of these vaccine strains need to be evaluated for a better control of the IBD in Senegal.
In conclusion, we have studied the genetic evolution of the Senegalese field IBDVs isolated in 1979, 1999, 2007, 2012, 2013 and 2014. Thus, on the basis of nucleotide and deduced amino acid sequences, the study reveals that the majority of viruses circulating in Senegal is (VV) IBDV and indicated (VV) IBDV as a main cause of substantial economic losses in the Senegalese poultry industry. We found that all Senegalese (VV) IBDVs cluster phylogenetically with (VV) IBDVs from Nigeria in an African genetic lineage, independent of the Asian/European genetic lineage. The origins of the African genetic lineage remain unclear, and need to be investigated. The study indicates genetic stability over time within the VP2 variable domain since 1979.
The authors have not declared any conflict of interests.
REFERENCES
|
Adamu J, Owoade AA, Abdu PA, Kazeem HM, Fatihu MY (2013). Characterization of field and vaccine infectious bursal disease viruses from Nigeria revealing possible virulence and regional markers in the VP2 minor hydrophilic peaks. Avian Pathol. 42:420-433.
Crossref
|
|
|
|
Bayliss CD, Spies U, Shaw K, Peters RW, Papageorgiou A, Műller H, Boursnell MEG (1990). A comparison of the sequences of segment A of four Infectious bursal disease virus strains and identification of a variable region in VP2. Gen. Virol. 71:1303-1312.
Crossref
|
|
|
|
|
Brandt M, Yao K, Liu M, Heckert RA, Vakharia VN (2001). Molecular determinants of virulence, cell tropism, and pathogenic phenotype of infectious bursal disease virus. J. Virol. 75:11974-82.
Crossref
|
|
|
|
|
Dormitorio TV, Giambrone JJ, Duck LW (1997). Sequence comparisons of the variable VP2 region of eight infectious bursal disease virus isolates. Avian Dis. 41:36-44.
Crossref
|
|
|
|
|
Islam MA, Khatun MM, Rahman MM, Hossain MT (2005). Serologic and pathogenic characterization of infectious bursal disease virus isolated from broiler chickens Bangladesh. Veterinarian 22(2):57-64.
|
|
|
|
|
Ismail NM, Saif YM, Moorhead PD (1998). Lack of pathogenicity of five serotype 2 infectious bursal disease viruses in chickens. Avian Dis. 32:757-759.
Crossref
|
|
|
|
|
Jackwood DJ, Sommer-Wagner SE, Crossley BM, Stoute ST, Woolcock PR, Charlton BR (2011). Identification and pathogenicity of a natural reassortant between a very virulent serotype 1 infectious bursal disease virus (IBDV) and a serotype 2 IBDV. Virology 420:98-105.
Crossref
|
|
|
|
|
Kasanga C J, Yamaguchi T, Wambura PN, Maeda- Machang'u AD, Ohya K, Fukushi H (2007). Molecular characterization of infectious bursal disease virus (IBDV): diversity of very virulent IBDV in Tanzania. Arch. Virol. 152:783-790.
Crossref
|
|
|
|
|
Kasanga C J, Yamaguchi T, Munangandu HM, Ohya K, Fukushi H (2013). Molecular epidemiology of infectious bursal disease virus in Zambia. J. South Afr. Vet. Assoc. 84(1):1-4.
Crossref
|
|
|
|
|
Martin AM, Fallacara F, Barbieri I, Tosi G, Rivallan G, Eterradossi N, Ceruti R, Cordioli P (2007). Genetic and antigenic characterization of infectious bursal disease viruses isolated in Italy during the period 2002-2005. Avian Dis. 51:863-872.
Crossref
|
|
|
|
|
Mawgod, SA, Arafa, AS, Hussein, HA (2014). Molecular genotyping of the infectious bursal disease virus (IBDV) isolated from Broiler Flocks in Egypt. Int. J. Vet. Sci. Med. 2:46-52.
Crossref
|
|
|
|
|
Műller H, Islam MR, Rauea R (2003). Research on infectious bursal disease-the past, the present and the future. Vet. Microbiol. 97:153-165.
Crossref
|
|
|
|
|
Owoade AA, Mulders MN, Kohnen J, Ammerlaan W, Muller CP (2004). High sequence diversity in infectious bursal disease virus serotype 1 in poultry and turkey suggests West-African origin of very virulent strains. Arch. Virol. 149:653-672.
Crossref
|
|
|
|
|
Sagna F (1975). Preliminary note on the appearance of a new avian disease in Senegal: Gumboro disease. Senegalese Institute of Agricultural Research (ISRA), National Laboratory for Livestock and Veterinary Research (LNERV).
|
|
|
|
|
Snyder DB, Vakharia VN, Savage PK (1992). Naturally occuring-neutralizing monoclonal antibody escape variants define the epidemiology of infectious bursal disease viruses in the United States. Arch. Virol. 127:89-101.
Crossref
|
|
|
|
|
Tamura K, Daniel P, Nicholas P, Glen S, Masatoshi N, Sudhir K (2011). MEGA5: Molecular Evolutionary Genetics Analysis Using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony. Method Mol. Biol. Evol. 28:2731-2739.
Crossref
|
|
|
|
|
Van den Berg TP (2000). Acute infectious bursa disease in poultry: a review. Avian Pathol. 29:175-194.
Crossref
|
|
|
|
|
Vakharia VN, He J, Ahamed B, Snyder DB (1994). Molecular basis of antigenic variation in infectious bursal disease virus. Virus Res. 31:265-273.
Crossref
|
|
|
|
|
Zierenberg K, Nieper H, Van den Berg TP, Ezeokoli CD, Voss M, Müller H (2000). The VP2 variable region of African and German isolates of infectious bursal disease virus: comparison with very virulent, classical virulent and attenuated tissue culture-adapted strains. Arch. Virol.145:113-125.
Crossref
|
|