ABSTRACT
The aim of the present research was to evaluate the use of the sugary Kefir grains as a starter culture for new rice cereal-based Kefir beverage. Fermentation was performed by inoculating Kefir grains in rice extract. Flasks containing Kefir grains and substrates were statically incubated at 28°C for 24 h. The microbiota of sugary Kefir grains and rice cereal-based Kefir beverage were genera Lactobacillus, Lactococcus and Acetobacter as well as yeasts, such as Saccharomyces, Kluyveromyces, Lachancea and Kazachstania. The sugary Kefir grains were able to ferment the rice extract and produced Kefir beverage that are functional and healthy that satisfy nutrition-related conditions such as allergies and malabsorption, food intolerances, and lifestyle choices, for example vegetarianism and low salt. The use of starter cultures as Kefir grains offers a promising tool for innovation and diversification of cereal-based beverages. This study was the first to report the rice cereal-based Kefir beverage production. This result opens up perspectives for this innovative application of sugary Kefir grains for developing cereal-based beverages.
Key words: Probiotic, fermentation, food intolerances, vegetarianism.
The use of cereals grains around 10,000 B.C. led to cereals becoming source of nutrients for population throughout the world. Cereals grains are a source of dietary fibers and carbohydrate and provide nutrients such as vitamins and minerals and low-gluten or gluten-free. Soy-based food and beverages, cereals, fruits, and vegetables have been considered as ingredients for functional food/beverages that satisfy dietary lifestyles such as allergen-free and veganism (Peyer et al., 2016; Fiorda et al., 2017). On a worldwide basis, rice is the prevailing crops in terms of area reserved for cereal cultivation and total cereal production. In Segundo, the world rice production in 2017 was 756.7 million tonnes. However, cereals, such as quinoa and buckwheat, havegenerated interest in western countries, because of their higher content in dietary fiber, starch, vitamins and minerals (Peyer et al., 2016). Kefir is a traditional Middle Eastern beverage. It originated in the Caucasus in Asia in thousands of years (Magalhães et al., 2010a; Nalbantoglu et al., 2014; Fiorda et al., 2017). Kefir is a symbiosis between yeast and bacteria.
A vast variety of different species of microorganisms formed the Kefir grains (Miguel et al., 2011; Nalbantoglu et al., 2014; Viana et al., 2017; Magalhães-Guedes et al., 2017; Fiorda et al., 2017; Roos and Vuyst, 2018). Lactobacillus genera are the most frequent in kefir. Other lactic bacteria including Lactococcus and Leuconostoc genera are also common in Kefir (Nalbantoglu et al., 2014; Magalhães-Guedes et al., 2017; Fiorda et al., 2017; Roos and Vuyst, 2018). Acetobacter genera represent acetic bacteria and the yeast isolates are Kluyveromyces, Candida and Saccharomyces genera (Leite et al., 2013; Marsh et al., 2013; Viana et al., 2017). The microbial species from Kefir grains carried out three types of fermentation during the process: lactic, alcoholic and acetic. Kefir can easily adapt to different substrates and lead to production of new probiotic products (Nalbantoglu et al., 2014; Fiorda et al., 2016; Fiorda et al., 2017).
“Traditional” way of Kefir beverage production is using pasteurized or Ultra-high-temperature (UHT) processing treated milk. Kefir beverage is mainly considered a probiotic resource (Leite et al., 2013). Kefir may help bridge the gap between the health benefits and consumption of non-dairy foods and provide the benefits of probiotic without milk or diary product consumption (Leite et al., 2013; Garofalo et al., 2015; Fiorda et al., 2016). Due to the numerous positive effects of Kefir on the human health, alternative substrates others may be used for kefir grains fermentation. Names of the resulting beverages are changed in case additional fruit, molasses or vegetable are used as medium of fermentation (Magalhães et al., 2010a,b; Miguel et al., 2011; Nalbantoglu et al., 2014; Fiorda et al., 2016, 2017). The adaptation of Kefir grains into different substrates has shown potential for production of Kefir beverages with distinct sensory characteristics and functional proprieties. For all the aforementioned, the objective of this study was the use of Kefir grains as starter culture for fermented rice cereal-based beverage.
The raw materials used were polished rice type 1 (Oryza sativa) and brown sugar, from the city of Salvador, Bahia. The sugary Kefir grains were obtained from the Probiotics Laboratory of the Federal University of Bahia, UFBA.
Preparation of rice extract
The rice extract was elaborated based on the methodology proposed by Peyer et al. (2016). The Kefir grains were initially washed in water to reduce or eliminate soil contamination. In a stainless, the beans were cooked in the ratio of 1 part grain to 2 part water (1: 2 w/w), for 30 min. The product was filtered, drained and homogenized.
Experimentation and chemical analysis
Samples (in triplicate), control and rice extract, were inoculated with sugary Kefir grains following the traditional method (Figure 1). The control (C) and test (T) samples were 1000 mL of filtered 5% sugary water and rice extract, respectively. The proportion of 10% sugary Kefir grains was added in sterile glass containers (5% sugary water and rice extract). The fermentation time of the samples corresponded to 24 h/28°C. pH, acidity, and Kefir grains weight mass, were performed in 0 and 24 h. pH was determined using a Neomed model pH meter. Titratable acidity (in lactic acid) and mass weight (Shimadzu) were done according to the rules of the Adolfo Lutz Institute (2008).
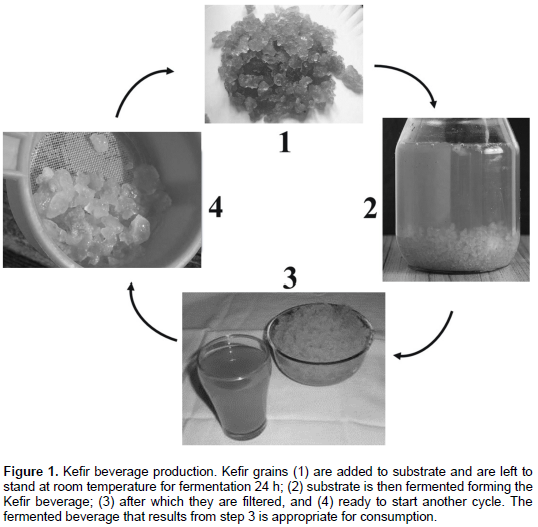
DNA extraction and PCR-DGGE analysis
The sugary Kefir (grains and rice cereal-based Kefir beverage) microbiological analysis was carried out at the Molecular Biology Laboratory of the Federal University of Lavras, UFLA, Brazil. For analysis, 1 g of sugary Kefir grains and 1 mL of rice cereal-based Kefir beverage sample were transferred into a plastic tube and was subjected to DNA extraction using a Nucle-oSpin Tissue kit (Macherey-Nagel, Du ¨ren, Germany). DNA extraction was performed according to the manufacturing instructions. The bacterial community DNA was ampliï¬ed with primers 338fgc and 518r spanning the V3 region of the 16S rDNA gene (Puerari et al., 2015). The yeast community DNA was ampliï¬ed using the primers NS3 and YM951r (Magalhães et al., 2010b). The ampliï¬cation was carried according to the method of Magalhães et al. (2010b). Polymerase chain reaction (PCR) products were analysed by denaturing gradient gel electrophoresis (DGGE) using a Bio-Rad DCode Universal Mutation Detection System (Bio-Rad, Richmond, CA, USA). Samples were applied to 8% (w/v) polyacrylamide gels in 0.5% TAE.
Optimal separation was achieved with a 15 to 55% urea-formamide denaturing gradient for the bacterial community and a 12 to 50% gradient for the yeast community, where 100% is deï¬ned as 7 M urea and 40% (v/v) formamide. Electrophoresis was carried out for 3 h at 200 V at 60°C, and the gels were stained with SYBR-Green I (Molecular Probes, Eugene, OR, USA) (1:10,000 v/v) for 30 min. The gels were visualised via U. V. transilluminator, and images were captured using a Polaroid camera (Concord, USA). The bands were excised with a sterile surgical blade and stored at -20°C until further analysis. DGGE bands were excised from the acrylamide gels and the fragments were puriï¬ed using the QIAEX II gel extraction kit (Qiagen, Chatsworth, CA, USA). DNA recovered from each DGGE band was reampliï¬ed using the primers 338f (without GC clamp) and 518r for bacteria and NS3 (without GC clamp) and YM951r for yeast. The PCR amplicons were then sequenced (Applied Biosystems, Foster City, CA, USA). GenBank searches (http://www. ncbi.nlm.nih.gov/BLAST/) were performed to determine the closest known relatives of the partial ribosomal DNA sequences obtained.
Statistical analysis
In order to obtain the means of the control and test samples, Statistical package for Social Science (SPSS) statistical software, version 20.0 was used. Student's t test, using the 95% confidence level, was also used.
After 24 h fermentation period, a significant difference (p<0.05) was observed between the Kefir grains weight of the control (C) and test (T) samples (Table 1). The cell mass (T) growth was higher in relation to the (C) cultures (Table 1). According to Magalhães et al. (2010a, b), the amount of Kefiran (polysaccharide) released during the fermentation depends on the microorganisms involved, the culture medium composition, the temperature and the fermentation time. Probably, the growth of the (T) samples is related to the greater availability of nutrients present in the rice extract. The decreased pH value (~4.3 to ~3.5) and the increased acidity (~0.2 to ~0.3) during 24 h of fermentation are shown in Table 2. These observations indicated that the fermentation process was followed by the production of acids. This also demonstrates that the sugary Kefir grains were able to ferment the rice extract. The low pH value (~3.5) and high acidity value (~0.3) present at the end of fermentation appears to be responsible for the presence of lactic acid bacteria as the major bacterial species at 24 h (Table 2). Traditionally, many plating procedures are only partially selective and exclude members of the microbial community (Miguel et al., 2011; Nalbantoglu et al., 2014; Corona et al., 2016; Fiorda et al., 2017; Cho et al., 2018; Roos and Vuyst, 2018).
Thus, to determinate the total composition of microbiota in the sugary Kefir grains and rice cereal-based Kefir beverage, PCR-DGGE analysis was used. The V3 region of the 16S rDNA gene of the bacteria and NS3 region of the 18S rDNA gene of the yeast were ampliï¬ed and representative DGGE ï¬ngerprints are as shown in Figure 2. To determine the composition of microbiota, individual bands observed in the PCR-DGGE proï¬les were excised from the acrylamide gel and re-ampliï¬ed to provide a template for sequencing. After BLAST analysis, sequence results showed between 99 and 100% identity with the sequences retrieved from GenBank accession numbers. DGGE bands a and d were clearly identiï¬ed as Lactobacillus paracasei, Lactobacillus parabuchneri, Lactobacillus keï¬ri, Lactococcus lactis, Lactobacillus casei, L. paracasei subsp. paracasei, Leuconostoc citreum, L. paracasei subsp. tolerans, Lactobacillus buchneri, Acetobacter lovaniensis, Saccharomyces cerevisiae, Kluyveromyces lactis, Lachancea meyersii and Kazachstania aerobia. Descriptions of the different types of yeast and bacteria present in different Kefir grains have been provided by different authors (Jianzhong et al., 2009; Magalhães et al., 2010a; Miguel et al., 2011; Nalbantoglu et al., 2014; Viana et al., 2017; Corona et al., 2016; Fiorda et al., 2017; Cho et al., 2018). Previous results have shown that three groups of microorganisms co-exist in Kefir grains: lactic acid bacteria, acetic acid bacteria and yeast (Jianzhong et al., 2009; Miguel et al., 2011; Nalbantoglu et al., 2014; Viana et al., 2017; Corona et al., 2016; Fiorda et al., 2017; Cho et al., 2018; Roos and Vuyst, 2018).
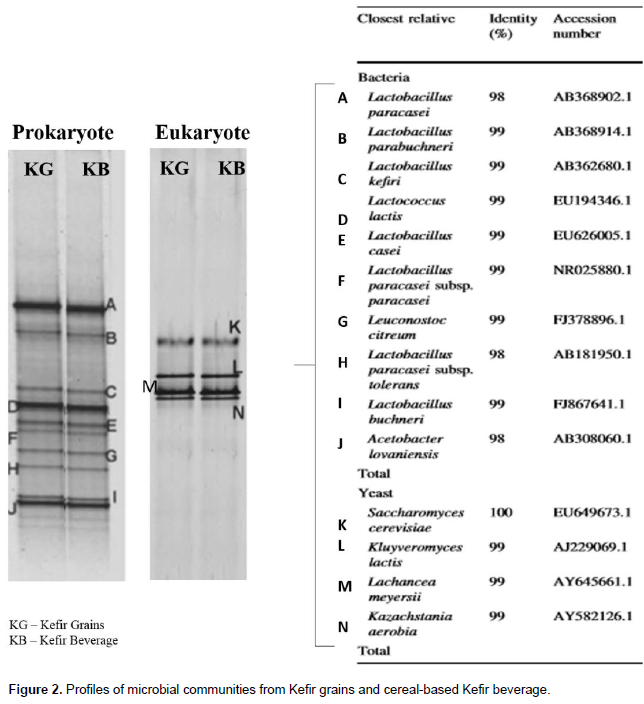
This data indicated that the sugary Kefir grains contained a diverse spectrum lactic acid bacteria group including Lactobacillus, Lactococcus and Leuconostoc. Another important bacterium found in sugary Kefir grains is L. keï¬ri. There are reports on the presence of L. keï¬ri as a member of the lactic acid microbiota in Keï¬r grains (Garrote et al., 2001; Jianzhong et al., 2009; Corona et al., 2016; Fiorda et al., 2017; Cho et al., 2018; Roos and Vuyst, 2018). The following lactic acid bacteria were also found: L. parabuchneri, L. lactis and L. casei, besides the species of L. citreum. Previous studies showed that a variety of lactic acid bacteria’s different species have been isolated and identiï¬ed in Kefir grains from around the world (Jianzhong et al., 2009; Magalhães et al., 2010a; Miguel et al., 2011; Nalbantoglu et al., 2014, Corona et al., 2016; Fiorda et al., 2017; Cho et al., 2018; Roos and Vuyst, 2018). The acetic acid species, A. lovaniensis, was also identiï¬ed. The species Acetobacter pasteurianus has been also described in fermented Kefir beverages (Corona et al., 2016; Viana et al., 2017; Fiorda et al., 2017; Cho et al., 2018). The lactose-fermenting yeast, K. lactis was identified in the sugary Kefir grains together with non-lactose-fermenting yeast S. cerevisiae, K. aerobia and L. meyersii (Figure 2). Of these yeasts, S. cerevisiae represented the most commonly identiï¬ed yeast isolates in Kefir grains. The presence of S. cerevisiae contributes to the organoleptic quality enhancement of the Keï¬r beverages, promoting a refreshing and pungent taste (Cho et al., 2018). The results demonstrated that rice extract could be an ideal alternative substrate for the production of functional cultured Kefir beverage, especially for vegans and lactose intolerant consumers, because the microorganisms of the Kefir grains successfully fermented the rice extract.
The results indicate that genera of bacteria, such as Lactobacillus, Lactococcus and Acetobacter, as well as yeast, such as Saccharomyces, Kluyveromyces, Lachancea and Kazachstania were the microorganisms present in sugary Kefir grains and rice cereal-based Kefir beverage. The sugary Kefir grains were able to ferment the rice extract and produce Kefir beverage functional and healthy that satisfy nutrition-related conditions such as allergies and malabsorption, food intolerances, and lifestyle choices, for example vegetarianism and low salt. This study is the first to report the rice cereal-based Kefir beverage production. The use of starter cultures as Kefir grains offers a promising tool for innovation and diversification of cereal-based beverages.
The authors have not declared any conflict of interests.
REFERENCES
|
Corona O, Randazzo W, Miceli A, Guarcello R, Francesca N, Esten H, Moschetti G, Settanni L (2016). Characterization of kefir-like beverages produced from vegetable juices. LWT-Food Sci. Technol. 66:572-581.
|
|
|
|
Fiorda FA, de Melo Pereira GV, Thomaz-Soccol V, Rakshit SK, Pagnoncelli MGB, Vandenberghe LPS, Soccol CR (2017). Microbiological, biochemical, and functional aspects of sugary kefir fermentation - A review. Food Microbiol. 66:86-95.
Crossref
|
|
|
|
|
Fiorda FA, de Melo Pereira GV, Thomaz-Soccol V, Medeiros AP, Rakshit SK, Soccol CR (2016). Development of kefir-based probiotic beverages with DNA protection and antioxidant activities using soybean hydrolyzed extract, colostrum and honey. LWT- Food Sci. Technol. 68:690-697.
|
|
|
|
|
Garofalo C, Osimani A, Milanovic V, Aquilanti L, De Filippis F, Stellato G, Di Mauro S, Turchetti B, Buzzini P, Ercolini D, Clementi F (2015). Bacteria and yeast microbiota in milk kefir grains from different Italian regions. Food Microbiol. 49:123-133.
Crossref
|
|
|
|
|
Garrote GL, Abraham AG, De Antoni GL (2001). Chemical and microbiological characterization of keï¬r grains. J. Dairy Res. 68:639-652.
Crossref
|
|
|
|
|
Instituto Adolfo Lutz (São Paulo) (2008). Métodos físico-químicos para análise de alimentos /coordenadores Odair Zenebon, Neus Sadocco Pascuet e Paulo Tiglea. São Paulo: Instituto Adolfo Lutz, 1020p.
|
|
|
|
|
Jianzhong Z, Xiaoli L, Hanhu J, Mingsheng D (2009). Analysis of the microflora in Tibetan keï¬r grains using denaturing gradient gel electrophoresis. Food Microbiol. 26:770-775.
Crossref
|
|
|
|
|
Leite AMO, Miguel MAL, Peixoto RS, Rosado AS, Silva JT, Paschoalin VMF (2013). Microbiological, technological and therapeutic properties of kefir: a natural probiotic beverage. Braz. J. Microbiol. 44:341-349.
Crossref
|
|
|
|
|
Magalhães KT, Dias D, Pereira GVM, Oliveira JM, Domingues L, Teixeira JA, Almeida e Silva JB, Schwan RF (2011a). Chemical composition and sensory analysis of cheese whey-based beverages using kefir grains as starter culture. Inter. J. Food Sci. Technol. 46:871-878.
Crossref
|
|
|
|
|
Magalhães KT, Dragone G Pereira GVM, Oliveira JM, Domingues L, Teixeira JA, Almeida e Silva JB, Schwan RF (2011b). Comparative study of the biochemical changes and volatile compounds during the production of novel whey-based kefir beverages and traditional milk kefir. Food Chem. 126:249-253.
Crossref
|
|
|
|
|
Magalhães KT, Pereira GVM, Campos CR, Dragone G, Schwan RF (2011c). Brazilian kefir: structure microbial communities and chemical composition. Braz. J. Microbiol. 42:693-702.
Crossref
|
|
|
|
|
Magalhães KT, Pereira GVM., Dias DR, Schwan RF (2010a). Microbial communities and chemical changes during fermentation of sugary Brazilian kefir. World J. Microbiol. Biotechnol. 26:1241-1250.
Crossref
|
|
|
|
|
Magalhães KT, Pereira MA, Nicolau A, Dragone G, Domingues L, Teixeira JA, Silva JBA., Schwan R F (2010b). Production of fermented cheese whey-based beverage using keï¬r grains as starter culture: evaluation of morphological and microbial variations. Bioresour. Technol. 101:8843-8850.
Crossref
|
|
|
|
|
Magalhães-Guedes KT, Ferreira CD, Magalhães KT, Schwan RF, Costa JAV, Nunes IL (2017). Microorganisms in functional food supplementation: A review. Afr. J. Microbiol. Res. 8:319-326.
|
|
|
|
|
Marsh AJ, O'Sullivan O, Hill C, Ross RP, Cotter PD (2013). Sequencing-Based Analysis of the Bacterial and Fungal Composition of Kefir Grains and Milks from Multiple Sources. Plos One 8:1-11.
Crossref
|
|
|
|
|
Miguel MGCP, Cardoso PG, Magalhães KT, Schwan RF (2011). Profile of microbial communities present in tibico (sugary kefir) grains from different Brazilian states. World J. Microbiol. Biotechnol. 27:1875-1884.
Crossref
|
|
|
|
|
Nalbantoglu U, Cakar A, Dogan H, Abaci N, Ustek D, Sayood K, Can H (2014). Metagenomic analysis of the microbial community in kefir grains. Food Microbiol. 41:42-51.
Crossref
|
|
|
|
|
Peyer LC, Zannini E, Arendt E (2016). Lactic acid bacteria as sensory biomodulators for fermented cereal-based beverages. Trends Food Sci. Technol. 54:17-25.
Crossref
|
|
|
|
|
Puerari C, Magalhães-Guedes KT, Schwan RF (2015). Physicochemical and microbiological characterization of chicha, a rice-based fermented beverage produced by Umutina Brazilian Amerindians. Food Microbiol. 46:210-217.
Crossref
|
|
|
|
|
Roos J, Vuyst L (2018). Acetic acid bacteria in fermented foods and beverages. Curr Opin. Biotechnol. 49:115-119.
Crossref
|
|
|
|
|
Viana RO, Magalhães-Guedes KT, Braga Jr. RA, Dias, DR, Schwan RF (2017). Fermentation process for production of apple-based kefir vinegar: Microbiological, chemical and sensory analysis. Braz. J. Microbiol. 48:592-601.
Crossref
|
|