ABSTRACT
Plastic polymers are petroleum-derived synthetic materials that have multiple uses in everyday life, but their excessive production has led to the accumulation of approximately 1,000 million tons of residues, causing negative ecological impacts. This study analyzed the biological degradation in liquid medium of polyurethane, polystyrene, and polyethylene samples by filamentous fungi isolated from Antarctica. The plastic samples were used without pretreatment or pretreated with an artificial aging UV chamber according to ASTM G155 for 500 h, inoculated or not with the Antarctic fungi (Penicillium, Geomyces, Mortierella species). Samples were incubated at 18°C for 90 days to determine potential fungal biodegradation. The physical-chemical and biological degradation of plastics were evaluated by analyzing the weight loss in function of time, and by determining possible changes in the chemical structure, using the technique of Fourier Transform Infrared Spectroscopy (FTIR). The polymers exposed to the artificial aging chamber resulted in the oxidative degradation of plastics (detected by morphological and structural changes), which favored their biodegradation. Out of the three fungal strains, Penicillium spp. presented the highest degradation percentage in aged plastics corresponding to 28.3% in polyurethane, and to 8.39 and 3.53% in polystyrene and low-density polyethylene, respectively.
Key words: Plastic aging, polymers, filamentous fungi, fungal biodegradation, deterioration.
Plastic polymers derived from fossil petroleum sources are used in the manufacture of short time disposable products which represent one of the main components of solid wastes. The United Nations Environment Program (UNEP) reported around 13 million tons of plastics that are dumped into the oceans annually. America, Japan and the European Union are the largest producers of this type of wastes. Besides, only 9% of these plastic wastes produced worldwide is recycled (ONU, 2018).
The problem caused by plastic waste can be solved by adding pro-oxidants or biologically degradable polymers during manufacturing, then, allowing their deterioration in the environment in less time (Chiellini et al., 2007). Another alternative is related to the use of microorganisms able to biodegrade some plastic polymers effectively (Bonhomme et al., 2003; Hermann et al., 2011). Biodegradation is a natural decomposition process of a substance or product by the action of biological agents, achieving its elimination or transformation to less dangerous products for nature (Arutchelvi et al., 2008). Moreover, plastic deterioration is related to the conditions of exposition. Environmental factors such as light, heat and/or biological activity induce changes in the functional properties of polymers, then, causing the rupture of bonds and chemical transformations. These changes are observed due to the formation of cracks and discoloration. Furthermore, solar radiation is one of the most harmful abiotic factors for polymers (Aradilla et al., 2007; Paço et al., 2017). Scientific reports indicate that the biological degradation of polymers depends on the polymer characteristics, the nature of the applied pretreatments, the polymer surface area, the type and microbial activity, as well as temperature, humidity, and nutrient availability (Bonhomme et al., 2003; Wu et al., 2017).
Researchers from Yale University reported the microscopic endophytic fungus Pestaloptiosis species at the Yasuni National Park (Ecuador), and indicated that this fungus degrades polyurethane (PU) under aerobic and anaerobic conditions (Russell, 2011). Different polymer-degrading microorganisms have been described; for instance, Koutny et al. (2009) indicated that Rhodococcus species strains may form a biofilm on a low density polyethylene sheet (LDPE), suggesting potential assimilation into microorganisms. On the other hand, Roy et al. (2008) mentioned that LDPE films were biodegradable by bacteria of the genus Bacillus. Other studies were also reported about the deterioration of the surface of polyethylene films due to fungal activity. Moreover, Ojeda et al. (2009) and Zahra et al. (2010) agree that the genera Aspergillus and Penicillium species, are capable of degrading polyethylene.
Thus, both fungi and bacteria are the most utilized microbial groups for biodegrading synthetic polymers. However, many microbial species have not yet been evaluated for such environmental purposes, resulting in limited available information regarding plastic-degrading microorganisms, and the involved enzymes on it. For this reason, this work performed an experimental bioassay for determining the degradation process of polyurethane, polystyrene, and polyethylene, by inoculating three strains of filamentous fungi, isolated from Antarctica.
Biological sampling site
Fungal strains were isolated from Antarctic soils by researchers from the Biotechnology Research Center of Ecuador (CIBE) in conjunction with the Instituto Antártico Ecuatoriano (INAE) in charge of the Pedro Vicente Maldonado Scientific Station, located on Greenwich-Antarctica Island (South Shetland Islands, Antarctic Peninsula, 62° 26' 57" S, 9° 44' 27" W). This project began with 40 fungal isolates that are deposited at the microorganism bank of CIBE-ESPOL. The isolates were reactivated on Papa Dextrose Agar (PDA) culture medium, at a temperature of 18°C. Once reactivated, a preliminary fungal screening was carried out, where the capacity of the microorganisms to grow in the presence of the polymers was evaluated. After this preliminary evaluation, three fungal strains were selected for further utilization in the present research (Table 1).

Fungal strain identification
Samples of mycelium of each fungal strain grown on Petri dishes were taken and placed in 1.5 mL microtubes added with 350 µL of extraction buffer (200 mM Tris, 250 mM NaCl, 25 mM EDTA, 0.5% SDS, pH 8.5), and 150 µL of 3 M sodium acetate, pH 5.2 and placed at -20°C for 10 min. Microtubes were centrifuged at maximum speed (14000 rpm) for 10 min. Then, supernatant was carefully transferred in a new 1.5 mL microtube in which 500 µL of isopropanol was added and incubated for 5 min at room temperature, then, centrifuged for 2 min at maximum speed (14000 rpm). After centrifugation, the supernatant was removed and the DNA pellet was washed with 50 µL of 70% ethanol, followed by centrifugation for 2 min with the lid open to evaporate the ethanol. Finally, 30 µL of sterile ultrapure water was added to the pellet, and resuspended and stored at -20°C. The DNA quantification was carried out in a NANODROP 2000. The molecular identification was based on the amplification of the ITS1 and ITS4 rDNA sequences (Cenis, 1992), amplified by a polymerase chain reaction (PCR) in a thermocycler (Mastercycler Nexus Thermocycler) using oligonucleotides ITS1 and ITS4 (White et al., 1990).
Each PCR reaction was added with 0.5 μL of genomic DNA, 13 μL of GOTAQ, 0.5 μL of each oligonucleotide, and 11 μL of ultrapure water, resulting in a final volume reaction of 25.5 μL with the following parameters for PCR: 98°C for 1 min, 35 cycles of 98°C for 45 s, 59°C for 40 s, 72°C for 1 min, and a final cycle of 72°C for 3 min. The amplified products were applied in a 1.5% agarose gel loading 3 µL of the PCR product and 0.5 µL of the 6x loading buffer (invitrogen). A 1 kb DNA Ladder molecular weight marker was used. The gel was run at 95 V for 15 min and subsequently visualized in an ultraviolet (UV) light transilluminator. The amplified products of the different fungal isolates were purified by using the DNA Purification by Centrifugation kit according to the manufacturer's instructions. The purified and concentrated products (30 ng/μL) were sent to the company Macrogen (Seoul, Korea).
Once the fungal sequences were obtained, these were analyzed with the free software program FinchTV. This allows cutting and cleaning DNA chains. After, the BLAST program was applied (NCBI, Basic Local Alignment Search Tool, 1988) for searching homologs of DNA or proteins based on the alignment of local type sequences, looking for identity (≥ 99%) with type strains reported. The sequences obtained with greater similarity were aligned with the method muscle for multiple alignments of the MEGA program. The Neighbor-Joining method was used to construct the phylogenetic tree (Saitou, 1987) with bootstrap values of 1000 iterations.
Polymeric materials
Samples of synthetic polymers were prepared by using 10, 20, and 40 mg, of low-density polyethylene, polystyrene and polyurethane, respectively. Part of the experiment was carried out by subjecting the polymer samples to a QUV accelerated aging treatment in a chamber with Xenon lamps, Q-SUN Xenon Test Chamber Model Xe-3-HBS following the ASTM G155 standard, “Standard Practice for Operating Xenon Arc Light Apparatus for Exposure of Non-Metallic Materials” for 500 h. Aged and non-aged samples were previously subjected to a disinfection process by soaking them successively for a minute in a solution of 10% sodium hypochlorite (v/v), sterile water heated to 60°C, and 70% ethanol (v/v).
In vitro polymer degradation
The degradation under in vitro conditions was carried out in 150 mL glass vials containing 50 mL of mineral liquid medium composed of: (g/L) KH2PO4, 1.0 g; NaNO3, 2.0 g; MgSO4 (7H2O), 0.05 g; KCL, 0.007 g; FeSO4 (7H2O), 0.01 g; NH4CL, 0.01 g; and plysurf, 0.01 g (Ishii et al., 2008), and autoclaved at 120°C for 20 min. At laminar flow chamber, the disinfected polymers were placed in the flasks with the mineral medium and inoculated with 50 mg of the respective fungal strains. Subsequently, the flasks were placed in an orbital shaker at 110 rpm, 18°C, for 90 days.
Polymer FTIR characterization
A polymer degradation analysis was performed using a Fourier Transform Infrared Spectroscopy (FTIR). The analyzed region corresponded to an interval between 4000 and 1000 cm-1, in where the presence of functional groups such as the amino (-NH), carbonyl (-C=O), nitrile (-C = N), amide (R-CO-N2), etc., can be determined. The structural changes that may occur in plastics as a result of a biodegradation process (Shah et al., 2008), and may be detected by the evolution of the bands corresponding to the mentioned functional groups involved in polymer degradation (Stuart, 2005). These tests were carried out on a Perkin-Elmer model Spectrum 100 infrared light spectrometer.
Both the dry weight of the polymer sample, and its degradation by Fourier transform infrared spectroscopy (FTIR) were determined every 30 days. Polymer mass loss was measured by the following equation:
Mass loss (%) = [(Wo-Wt)/Wo] × 100
where "Wo" is the initial weight of the polymer sample and "Wt" is the weight of the polymer after treatment and "t" indicates the number of days that the polymer remained in the mineral solution.
Absorption tests were performed to measure the percentage of water absorbed by each polymer for determining its hydrophilic character. The samples were dried in an oven for 30 min at 120°C. The percentage of water absorption was determined by the following equation:
Water absorption percentage (%) = [(Wi-Wf)/Wo] × 100
where "Wi" is the wet material weight and "Wf" is the weight of the dry material.
Statistical analysis
Statistical analyses were carried out with the variance analysis method (ANOVA) at a significance level of 5% (α = 0.05). Data were subjected to the SPSS program where the Duncan procedure for the mean comparison test was applied at 5% of significance level (Montgomery and Hines, 1993).
Sequence analysis and fungal identification
Results of the fungal identification are as shown in Figure 1 and Table 1. The CIBE-7-23a strain showed greater closeness to Penicillium adametzioides (99% identity), the strain CIBE-2-32a was phylogenetically related to two species of the genus Mortierella, but with the closest species of Mortierella turficola (98% identity). Likewise, the CIBE-12.1-11 strain is related to Geomyces species (98% identity). A fungus of Glomus species (Glomeromycota) was used as an external group.
In vitro polymer degradation
Polyethylene
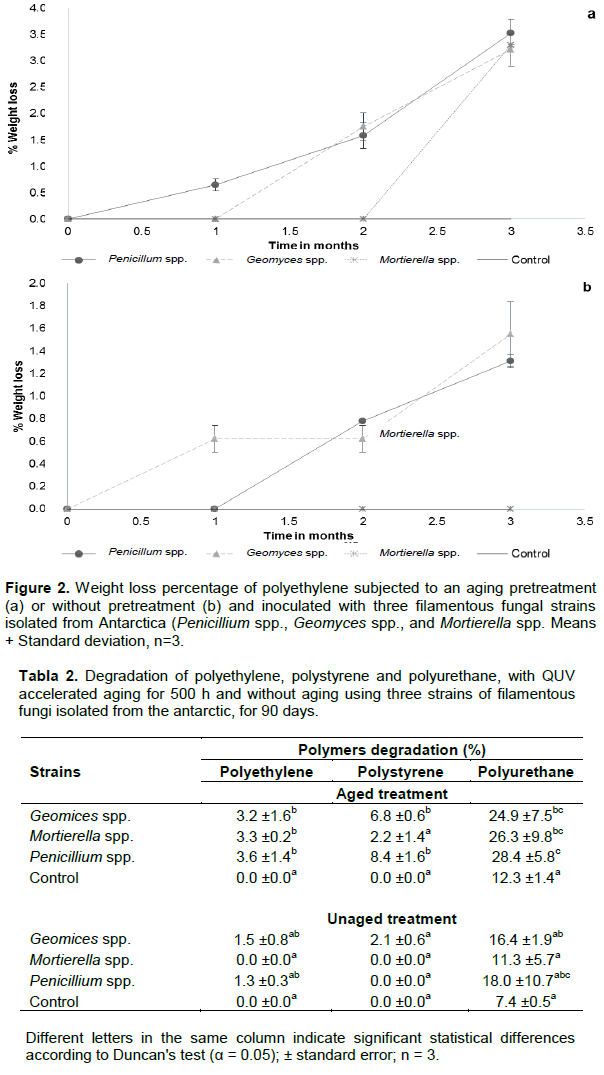
Figure 2 shows the degradation percentages for the low-density polyethylene (LDPE), after three months; samples pretreated with aging process and without aging inoculated with Penicillium spp. had a degradation of 3.5 ± 0.26% and 1.31 ± 0.06%, respectively (Figure 2a and b).

In case of Geomyces spp. inoculation, the degradation of both LDEP conditions was 3.22 ± 0.32 and 1.55 ± 0.29%, respectively, whereas by inoculating Mortierella species, the degradation values were of 3.31 ± 0.06 and 0%, respectively. Statistical analysis shows significant differences between the aging treatments and the control, while in the treatments without aging there are no significant differences between the treatments and the control (Table 2). The recorded low degradation percentages could be associated with the properties of this polymer such as high resistance to be attacked or modified by chemical agents, and to its high temperature (95°C) for inducing thermal degradation (Coreno and Mendez, 2010). Thus, it seems that high temperatures (40°C) may take an important effect on the biodegradation process of polymers (Kharoufeh, 2003), but in our experimental conditions, the temperature oscillated from 16 and 18°C. Moreover, this work focused on utilizing Antarctic fungal strains that are adapted to a lower temperature rather than those high temperatures used in other experimental reports (Kumari et al., 2009). Thus, the adaptation process of our fungal strains may be related to the low effectiveness on polymer degradation when compared with results obtained from other microorganisms isolated from Ecuador or elsewhere under different temperature conditions (Villa et al., 2009; Gajendiran et al., 2016). In all cases, results about the percentage of biodegradation were significantly higher than those achieved in this research. On another hand, the efficiency of a single fungal strain on polymer degradation may be lower than that from a fungal consortium. In this regards, Uribe et al. (2011) achieved a polyethylene biodegradation of 4.7 to 5.4% by using microbial consortia isolated from a landfill soil, for 60 days at 20°C. Moreover, Gajendiran et al. (2016) identified a strain of Aspergillus clavatus from landfill sites, which was able to degrade polyethylene. One of the main strategies for LDPE degradation is that mediated for microorganisms which may use this polymer as the sole source of carbon (Roy et al., 2008). Then, filamentous fungi such as Aspergillus and Penicillium have been reported as biodegraders of aged polyethylene films (Ojeda et al., 2009; Motta et al., 2009; Corti et al., 2010).
Polystyrene
Percentages of polystyrene (PS) degradation by Penicillium spp., Geomyces spp., and Mortierella are presented in Figure 3a and b. It is inferred that the fungal degradation was more effective on aged polymer than without aging, possibly due to the molecular alterations induced by the action of pretreatment with UV energy applied during 500 h. There were significant differences among treatments. The strains Penicillium and Geomyces spp. contributed on the polymer degradation of 8.39% ± 0.61 and 6.82% ± 0.87, respectively, while Mortierella spp. degraded this polymeric material by 2.19% ± 0.5. There were significant differences between aged and control treatments (Figure 3a and Table 2). Geomyces spp., was the only fungus able of producing a significant loss of mass of polystyrene for those samples without aging pretreatment, showing a maximum biodegradation (2.08% ± 0.21) after the second month of experimentation (Figure 3b). As accounted for polyethylene, the aging pretreatment made the polymer samples more susceptible for fungal attack and therefore, inducing a biodeterioration process. Some research reported different types of microorganisms able to deteriorate polystyrene, such as the worldwide distributed fungus Aureobasidium pullulans (Castiglia and Kuhar, 2015), known as "black yeast", that produces a large number of hydrolytic enzymes (Ma et al., 2007; Singh at al., 2008).
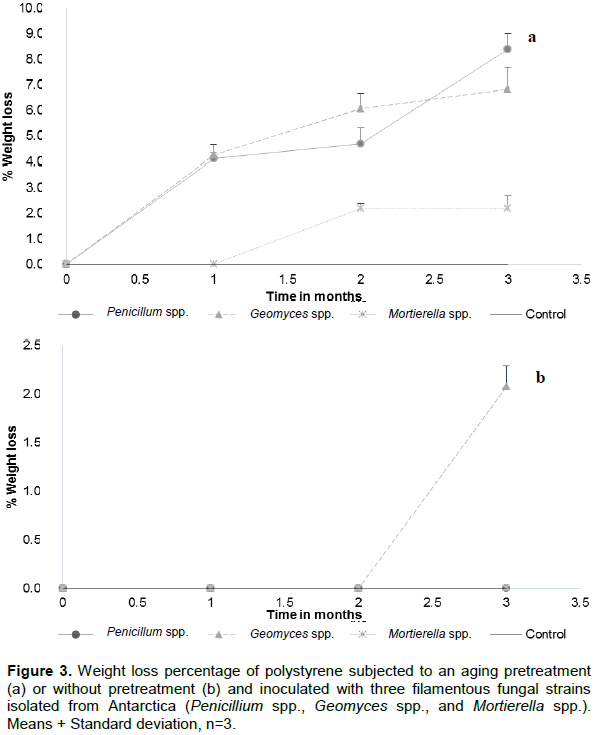
According to Castiglia and Kuhar (2015), A. pullulans is involved in the early stages of biodeterioration of plasticized materials like vinyl chloride and polystyrene. This fungus can be compared with our fungal strains for inducing the degradation of pretreated polymer since these fungi causes damage by mechanical action in where hyphae penetrate substrates that underwent an oxidation process (Webb et al., 2000). Furthermore, these results correlate with those obtained by FTIR, as described subsequently.
Polyurethane
Results of the polyurethane (PU) weight loss (%) at different sampling times are as shown in Figure 4a and b. The initial degradation rate of aged polyurethane was high due to the oxidation of water soluble oligomers (polymers of lower molecular weight that are hydrolyzed) present in the polymer matrix. On the other hand, the non-aging polyurethane has a monthly increase in the biodegradation process, but always smaller than that obtained for pretreated samples. Initially, the biodegradation occurs on the surface, since it is in contact with microorganisms, and the fungus spreads into the PU matrix (swelling process) (Amaral et al., 2012). The degradation mechanism of PU is achieved by fractions that are easy to transform and to hydrolyze since ester bonds are the first to break because they are more sensitive than urethane bonds. Moreover, the biodeterioration rate decreases because degradation of urethane bonds occurs at a slower rate (Zhang et al., 2009; Zhou et al., 2011).
Results from Figure 4 show that there are significant statistical differences among treatments (p <0.05). Pretreatment for aging polyurethane with the fungi after 3 months, produced a biodegradation around 26.5%; the most effective fungal strain was Penicillium spp. (28.34% ± 2.39). Taking into account the weight loss of the control sample (un-aged), the weight loss caused by fungal action was 16.09% (Figure 4a). All the aged treatments show significant differences with respect to the control (Tabla 2). In polyurethane without aging after 3 months, produced a biodegradation around 15.0%. The strain that degraded most effectively the polyurethane unaged was Penicillium spp. obtaining a 18.00% ± 10.7, but considering the results of the control sample, the effective loss per action of the fungi would be 10.7% (Figure 4b). In the case of the control sample, the polymer weight loss recorded with and without pretreatment was of 12.25% ± 1.40 and 7.35% ± 0.5, respectively. There are no statistically significant differences between the non-aging treatments and the control.
Scientific reports indicate that both bacteria and filamentous fungi are capable of degrading polyurethane; for instance, the fungal species like Geomyces pannorum, Nectria species, Penicillium inflatum, Plectosphaerella, Penicillium venetum, Neonectria ramulariae, and Penicillium viridicatum (Cosgrove et al., 2007); in all cases, the loss of mass would be related to the utilization of PU as a source of carbon or nitrogen (Urgun-Demirtas et al., 2007), and to the release of enzymes responsible of such degradation like proteases, esterases, and ureases (Ruiz et al., 1999). Most of the related experiments on PU degradation are focused on studies regarding bacteria. However, recent research has focused on the use of fungi (Amaral et al., 2012; Khan et al., 2017). Fungi are only capable of using organic carbon sources; thus, PU can be used as a potential carbon source for fungal growth (Loredo et al., 2017). Cosgrove et al. (2007) demonstrated in a soil microcosms that the sole application of yeast extract or its combination with Impranil, resulted in increased PU degradation up to 62%. Moreover, Loredo et al. (2017) applied three fungi (Trichoderma species, Aspergillus ustus, and Paecelomyces species) obtaining a 40% weight loss of rigid PU, after 30 days.
Analysis of the FTIR spectra of polymers
Polyethylene
The FTIR spectra obtained for LDPE films during the biodegradation process confirmed the existence of a possible microbial attack on polymers. The reduction of peaks corresponding to the C-H, C-O, and C = C groups would be related to polymer degradation. Figure 5a and b shows the spectra of the polyethylene samples used as control that, has not been subjected to the action of fungi. In these spectra, a series of characteristic peaks corresponding to the structure of the polyethylene can be observed. For pretreated polymer (Figure 5a), a wide absorption band appears between 1725 and 1700 cm-1 (centered around 1715 cm-1), indicating the modification of the polyethylene structure. The application of UV radiation, which has more energetic than visible radiation (> 298 kJ/mol), an oxidative polymer degradation occurred, generating the appearance of a band corresponding to the group C = O (Gulmine et al., 2003). Other reports indicate a decrease in the intensity of bands located at 719 cm-1, to the flexion of the CH group and 1472 cm-1, corresponding to the carbonyl group (-C = O) when the LDPE was in contact with bacteria (Das and Kumar, 2015). However, Figure 5a shows no considerable differences among spectra, by which it is difficult to assure biological action on this polymer related to the biodegradation. Nevertheless, the loss of weight achieved in the polymer could be due to the chemical action of either salts contained in the mineral medium or excretion or secretion of acids and pigments by fungi, which may modify the chemical properties and cause biodeterioration of polymers (Abrusci et al., 2009; Scott and Wiles, 2001).

Polystyrene
The spectra analysis showed that the material did not suffer a biodegradation process since no significant variations in the FTIRR spectra were observed in both control sample and samples subjected to fungal action. However, by analyzing the obtained spectra (Figure 5b and c) and the weight loss recorded for the different samples, it can be concluded that the material underwent a biodeterioration process due to the interaction of mycelium with the polymer. This interaction may exert a chemical action resulted for excretion or secretion of acids and pigments by fungi which are deposited on the support modifying its chemical properties (Gu, 2007; Abrusci et al., 2009).
Polyurethane
The UV aging treatment caused significant morphological and structural changes on polyurethane. This physical process generated an oxidative degradation of the polymer, which increased its susceptibility to microbial attack. Bands between 3500 and 3200 cm-1 are attributed to the overlap of the signal corresponding to the stretching and bending vibrations of bonds O-H, respectively (Jagtap et al., 2011; Spontón et al., 2013). The spectra of the control samples subjected or not to abiotic pretreatment (Figure 5e) show that aged samples had a widening of the band centered at 3300 cm-1, corresponding to the overlapping of the stretching of NH and OH groups. This widening would be related to the appearance of primary NH groups formed during the hydrolysis of urethane group that is benefited by the aforementioned pretreatment (Oprea and Oprea, 2016; Tavares and Schleder, 2016). The hydrolysis equation produced is as follows:
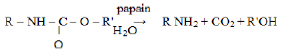

The FTIR spectra obtained for PU samples subjected to aging treatment, show differences in the area between 1720 and 1500 cm-1 that allow the comparison of the effect caused by the action of the three fungal strains and the sample used as control. Penicillium and Geomyces were the two strains that produced the greatest weight loss, the spectra show a signal around 1664 cm-1 corresponding to the amide group, and this signal is related to a hydrolysis process (Oprea and Oprea, 2016). The increase of the signal located at 1541 cm-1 in those samples subjected to the pretreatment in contact with the two mentioned fungi, corresponded to the vibration of the NH, which is probably a consequence of the aforementioned hydrolysis.
On the otherhand, by comparing the spectra of both graphs (Figure 5e and f) some displacements were found towards smaller numbers of waves of the peaks located in 1707.44 cm-1 for the aged control sample, and in 1707.36 cm-1 for the control sample without aging. These peaks are related to hydrogen bond interactions between NH groups and carbonyl groups (Loredo et al., 2017), as a consequence of a biodegradation action. In both spectra there were no variations in the area between 1000 and 600 cm-1 that corresponds to the polymer fingerprint (Vaghani et al., 2012).
Based on the evidence in this work, we can see that there are microorganisms in nature that have the ability to degrade certain wastes that are dangerous for the environment. A biotechnological approach based on the identification of the enzymes responsible for the degradation of these residues could make the degradation processes more efficient.
The present study evaluated the ability of three fungal strains to use LDPE, PS, and PU as the only carbon source. It was possible to identify that the aged materials are more susceptible to fungal attack in comparison to those without treatment. However, not all polymers were biodegraded as occurred for LDPE and PS that only accounted in a biodeterioration process. Unlike, the PU was susceptible to a biodegradation action, as demonstrated in the FTIR spectra. Thus, PU in the aging treatment was the most degraded polymer by the activity of the three fungal strains. The fungal strain that produced the highest degradation of polyurethane (28.34%), was Penicillium spp.
The authors have not declared any conflict of interests.
REFERENCES
|
Abrusci C, Marquina D, Santos A, Del Amo A, Corrales T, Catalina F (2009). Biodeterioration of cinematographic cellulose triacetate by Sphingomonas paucimobilis using indirect impedance and chemiluminescence techniques. International Biodeterioration Biodegradation 63(6):759-764.
Crossref
|
|
|
|
Amaral JS, Sepúlveda M, Cateto CA, Fernandes IP, Rodrigues AE, Belgacem MN, Barreiro MF (2012). Fungal degradation of lignin-based rigid polyurethane foams. Polymer Degradation and Stability 97(10):2069-2076.
Crossref
|
|
|
|
|
Aradilla D, Estrany F, Oliver R (2007). Degradación de residuos de materiales plásticos. Revista IngenieriÌa QuiÌmica 448(1):186-190.
|
|
|
|
|
Arutchelvi J, Sudhakar M, Arkatkar A, Doble M, Bhaduri S, Uppara PV (2008). Biodegradation of polyethylene and polypropylene. Indian Journal of Biotechnology 7(1):9-22.
|
|
|
|
|
Bonhomme S, Cuer A, Delort A. M, Lemaire J, Sancelme M, Scott G (2003). Environmental biodegradation of polyethylene. Polymer Degradation and Stability 81(3):441-452.
Crossref
|
|
|
|
|
Castiglia VC, Kuhar F (2015). Deterioration of expanded polystyrene caused by Aureobasidium pullulans var. melanogenum. Revista Argentina de Microbiologia 47(3):256-260.
Crossref
|
|
|
|
|
Coreno AJ, Méndez M (2010). Relación estructura-propiedades de polímeros. Revista Educación química de UNAM 21(4):291-299.
Crossref
|
|
|
|
|
Corti A, Muniyasamy S, Vitali M, Imam SH, Chiellini E (2010). Oxidation and biodegradation of polyethylene films containing pro-oxidant additives: Synergistic effects of sunlight exposure, thermal aging and fungal biodegradation. Polymer Degradation and Stability 95(6):1106-1114.
Crossref
|
|
|
|
|
Cosgrove L, McGeechan PL, Robson GD, Handley PS (2007). Fungal communities associated with degradation of polyester polyurethane in soil. Applied and Environmental Microbiology 73(18):5817-5824.
Crossref
|
|
|
|
|
Cenis JL (1992). Rapid extraction of fungal DNA for PCR amplification. Nucleic Acids Research 20(9):2380.
Crossref
|
|
|
|
|
Chiellini E, Corti A, D'Antone S (2007). Oxo-biodegradable full carbon backbone polymers - biodegradation behaviour of thermally oxidized polyethylene in an aqueous medium. Polymer Degradation and Stability 92(7):1378-1383.
Crossref
|
|
|
|
|
Das MP, Kumar S (2015). An approach to low-density polyethylene biodegradation by Bacillus amyloliquefaciens. 3 Biotech 5(1):81-86.
Crossref
|
|
|
|
|
Gajendiran A, Krishnamoorthy S, Abraham J (2016). Microbial degradation of low-density polyethylene (LDPE) by Aspergillus clavatus strain JASK1 isolated from landfill soil. 3 Biotech 6(1):52.
Crossref
|
|
|
|
|
Gu JD (2007). Microbial colonization of polymeric materials for space applications and mechanisms of biodeterioration: A review. International Biodeterioration Biodegradation 59(3):170-179.
Crossref
|
|
|
|
|
Gulmine JV, Janissek PR, Heise HM, Akcelrud L (2003). Degradation profile of polyethylene after artificial accelerated weathering. Polymer Degradation and Stability 79(3):385-397.
Crossref
|
|
|
|
|
Hermann B, Debeer L, Wilde B, Blok K, Patel M (2011). "To compost or no to compost: Carbon and energy footprints of biodegradable materials' waste treatment. Polymer Degradation and Stability 96:1159-1171.
Crossref
|
|
|
|
|
Ishii N, Inoue Y, Tagaya T, Mitomo H, Nagai D, Kasuya K (2008). Isolation and characterization of poly (butylene succinate)-degrading fungi. Polymer Degradation and Stability 93(5):883-888.
Crossref
|
|
|
|
|
Jagtap S, Yenkie MK, Das S, Rayalu S (2011). Synthesis and characterization of lanthanum impregnated chitosan flakes for fluoride removal in water. Desalination 273(2-3):267-275.
Crossref
|
|
|
|
|
Koutny M, Amato P, Muchova M, Ruzicka J, Delort AM (2009). Soil bacterial strains able to grow on the surface of oxidized polyethylene film containing prooxidant additives. International Biodeterioration Biodegradation 63(3):354-357.
Crossref
|
|
|
|
|
Khan S, Nadir S, Shah ZU, Shah AA, Karunarathna SC, Xu J, Hasan F (2017). Biodegradation of polyester polyurethane by Aspergillus tubingensis. Environmental Pollution 225:469-480.
Crossref
|
|
|
|
|
Kharoufeh JP (2003). Explicit results for wear processes in a Markovian environment. Operations Research Letters 31(3):237-244.
Crossref
|
|
|
|
|
Kumari K, Aanad RC, Narula N (2009). Microbial degradation of polyethylene (PE). The South Pacific Journal of Natural and Applied Sciences 27(1):66-70.
Crossref
|
|
|
|
|
Loredo A, Argüello A, Rodríguez-Herrera R, Gutiérrez-Sánchez G, Escamilla A, Aguilar C (2017). Biodegradacion fungica de poliuretano rígido. Química Nova 40(8):885-889.
|
|
|
|
|
Ma C, Ni X, Chi Z, Ma L, Gao L (2007). Purification and characterization of an alkaline protease from the marine yeast Aureobasidium pullulans for bioactive peptide production from different sources. Marine Biotechnology 9(3):343-351.
Crossref
|
|
|
|
|
Montgomery D, Hines W (1993). Probabilidad y estadística para ingeniería y administración. Management Science. Facultad de ciencias de la UNAM. Compañía Editorial: Continental S.A. México.
|
|
|
|
|
Motta O, Proto A, De Carlo F, De Caro F, Santoro E, Brunetti L, Capunzo M (2009). Utilization of chemically oxidized polystyrene as co-substrate by filamentous fungi. International Journal of Hygiene and Environmental Health 212(1):61-66.
Crossref
|
|
|
|
|
Paço A, Duarte K, da Costa JP, Santos P, Pereira R, Pereira ME, Freitas A, Duarte A, Rocha-Santos T (2017). Biodegradation of polyethylene microplastics by the marine fungus Zalerion maritimum. Science of the Total Environment 586:10-15.
Crossref
|
|
|
|
|
Ojeda TF, Dalmolin E, Forte MM, Jacques RJ, Tima FM, Vio FA (2009). Abiotic and biotic degradation of oxo-biodegradable polyethylenes. Polymer Degradation and Stability 94:965-970.
Crossref
|
|
|
|
|
Organización de las Naciones Unidas (ONU) (2018). O nos divorciamos del plástico, o nos olvidamos del planeta. (Blog) ONU Medio Ambiente/Shawn Heinrichs. ONU Noticias (30.01.2019). [accessed 2019 Sep 10].
View
|
|
|
|
|
Oprea S, Oprea V (2016). Biodegradation of crosslinked polyurethane acrylates/guar gum composites under natural soil burial conditions. e-Polymers 16(4):277-286.
Crossref
|
|
|
|
|
Roy PK, Titus S, Surekha P, Tulsi E, Deshmukh C, Rajagopal C (2008). Degradation of abiotically aged LDPE films containing pro-oxidant by bacterial consortium. Polymer Degradation and Stability 93(10):1917-1922.
Crossref
|
|
|
|
|
Ruiz C, Main T, Hilliard NP, Howard GT (1999). Purification and characterization of twopolyurethanase enzymes from Pseudomonas chlororaphis. International Biodeterioration Biodegradation 43(1-2):43-47.
Crossref
|
|
|
|
|
Russell J (2011). Biodegradation of Polyester Polyurethane by Endophytic Fungi. Applied and Environmental Microbiology 6076-6084.
Crossref
|
|
|
|
|
Saitou N (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molecular Biology and Evolution 4(4):406-425.
|
|
|
|
|
Scott G, Wiles DM (2001). Programmed-Life Plastics from Polyolefins: A New Look at Sustainability †. Biomacromolecules 2(3):615-622.
Crossref
|
|
|
|
|
Shah AA, Hasan F, Hameed A, Ahmed S (2008). Biological degradation of plastics: A comprehensive review. In Biotechnology Advances 26(3):246-265.
Crossref
|
|
|
|
|
Singh RS, Saini GK, Kennedy JF (2008). Pullulan: Microbial sources, production and applications. Carbohydrate Polymers 73(4):515-531.
Crossref
|
|
|
|
|
Spontón M, Casis N, Mazo P, Raud B, Simonetta A, Ríos L, Estenoz D (2013). Biodegradation study by Pseudomonas sp. of flexible polyurethane foams derived from castor oil. International Biodeterioration Biodegradation 85(5):85-94.
Crossref
|
|
|
|
|
Stuart B (2005). Infrared Spectroscopy. In Kirk-Othmer Encyclopedia of Chemical Technology (19). John Wiley & Sons, Inc.
Crossref
|
|
|
|
|
Tavares L, Schleder GR (2016). Bio-based polyurethane prepared from Kraft lignin and modified castor oil. Express Polymers Letter 10(11):927-940.
Crossref
|
|
|
|
|
Urgun-Demirtas M, Singh D, Pagilla K (2007). Laboratory investigation of biodegradability of a polyurethane foam under anaerobic conditions. Polymer Degradation and Stability 92(8):1599-1610.
Crossref
|
|
|
|
|
Uribe D, Giraldo D, Gutiérrez S, Merino F, Merino F (2011). Biodegradación de polietileno de baja densidad por acción de un consorcio microbiano aislado de un relleno sanitario, Lima, Perú. Revista Peruana de Biología 17(1):133-136.
Crossref
|
|
|
|
|
Vaghani SS, Patel MM, Satish CS (2012). Synthesis and characterization of pH-sensitive hydrogel composed of carboxymethyl chitosan for colon targeted delivery of ornidazole. Carbohydrate Research 347(1):76-82.
Crossref
|
|
|
|
|
Villa M, Rivera JD, Capilla V, Gardé JA (2009). Degradación biológica de polímeros mediante la selección y producción de potenciales cultivos iniciadores. Revista Técnica de Medio Ambiente 22(136):80-83.
|
|
|
|
|
Webb JS, Nixon M, Eastwood IM, Greenhalgh M, Robson GD, Handley PS (2000). Fungal colonization and biodeterioration of plasticized polyvinyl chloride. Applied and Environmental Microbiology 66(8):3194-3200.
Crossref
|
|
|
|
|
White TJ, Bruns T, Lee S, Taylor JW (1990). Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protocols: A Guide to Methods and Applications, pp. 315-322. Academic Pres Inn: Massachusetts.
Crossref
|
|
|
|
|
Wu W, Yang J, Criddle CS (2017). Estrategias de reducción y contaminación de microplásticos. Frontiers of Environmental Science and Engineering 11(6).
Crossref
|
|
|
|
|
Zhang Y, Xia Z, Huang H, Chen H (2009). A degradation study of waterborne polyurethane based on TDI. Polymer Testing 28(3):264-269.
Crossref
|
|
|
|
|
Zhou L, Yu L, Ding M, Li J, Tan H, Wang Z, Fu Q (2011). Synthesis and Characterization of pH-Sensitive Biodegradable Polyurethane for Potential Drug Delivery Applications. Macromolecules 44(4):857-864.
Crossref
|
|
|
|
|
Zahra S, Abbas SS, Mahsa MT, Mohsen N (2010). Biodegradation of low-density polyethylene (LDPE) by isolated fungi in solid waste medium. Waste Management 30(3):396-401.
Crossref
|
|