ABSTRACT
Pigments play an important role in physiological and molecular processes such as photosynthesis, survival to oxidative damage and resistance to ultra violet radiation in microorganisms. This study isolated and characterized pigment produced by Bacillus subtilis PD5 from soil sample of rice field. The pigment was extracted using three solvent system, namely, methanol, n-hexane and ethyl acetate. Fourier transform infrared spectroscopy (FTIR) technique was used for pigment characterization, while beta carotene was taken as standard. The molecular identification of bacterial strain was done by polymerase chain reaction (PCR) amplification of 16S rRNA gene sequence analysis. Sequence was submitted to National Center for Biotechnology Information (NCBI) database with accession number KY306687. Sequence analysis showed that the PD5 bacterial strain which produce red pigment is a novel strain of B. subtilis with 98 to 99% similarity. FTIR analysis of extracted red pigment showed similarity with beta carotene, indicating that the pigment is a derivative of carotenoid.
Key words: Bacillus subtilis PD5, 16S rRNA, Red pigment, Fourier transform infrared spectroscopy (FTIR).
Microbial cells accumulate pigments under certain culture conditions, with important industrial applications. Micro-organisms can serve as sources of carotenoids, the most widespread group of naturally occurring pigments. Colour is an important attribute for the consumer’s preference to buy foods. As a result various synthetic food colours have been manufactured but many of them comprise various hazardous effects (Fabre et al., 1993). The toxicity of synthetic colourants used as natural additives increases interest, therefore, emphasis on the production of bio-colour or natural colour extracted from fruits, vegetables, plant roots and microorganisms (Pattnaik et al., 1997). The industrial production of natural food colourants is already well-established and expanding. However, the use of natural colour is gaining attention. Production of microbial pigments can cut down the production cost of natural colours and it may be a cheaper source of natural food colourants. Hence, microbial pigment production is now one of the emerging fields of research which needs to be explored for commercial pigment production (Dufossé, 2009). Red biopigments produced as a secondary metabolite by some Serratia species (Serratia marcescens), actinomycetes and fungus Monascus species show antimicrobial activity and have a strong potential to develop antitumor drugs (Mekhael and Yousif 2009). The present study aimed at the screening of bacterial isolate capable of producing pigment in different organic solvents and of possible commercial importance. Promising pigment producing bacterial isolate was classified and identified through molecular characteristics. FTIR based characterisation of extracted pigment was done to find out chemical nature of pigment.
Isolation of bacteria
Pigment producing bacteria was isolated from the soil sample collected from the rice field where herbicide pendimethalin was supplemented for controlling weed. Soil sample (~1.0 kg) from 0 to 15 cm depth were taken and stored at 4°C. About 10 g soil was treated with triple recommended dose of crude pendimethalin at 30% Emulcified Concentration (EC) as it was used in the rice field and incubated for one week at room temperature for soil enrichment. After incubation, serial dilution, 10-1 to 10-8 was performed in sterile water and 10 μl from each dilution, was spread on nutrient agar plate. Plates were incubated at 28°C for 24 h. After incubation, five bacterial isolates PD1, PD2, PD3, PD4 and PD5 have been selected randomly, one bacterial isolate PD5 was found to produce pigment. The pigment producing bacterial isolate was used for pigment extraction.
Growth of bacterial isolate in mineral salt medium (MSM) having pendimethalin as carbon source
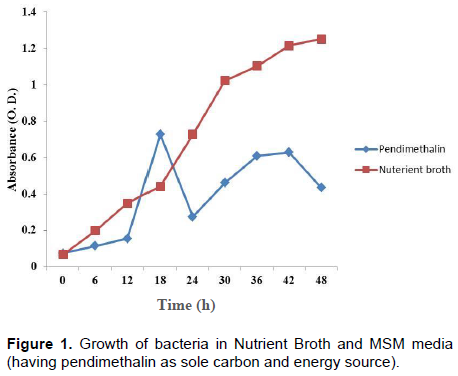
The bacterial isolate was allowed to grow in minimal salt media. Minimal medium as mineral salt liquid (MSL) and Nutrient Broth (NB), a complete medium were used throughout this study as described by Sambrook et al. (1989). Minimal salt media was supplemented with pendimethalin at a concentration of 50 mg/L. Single colony of bacteria was inoculated in 100 ml Nutrient broth and incubated at 28±2°C in incubator shaker at 120 rpm. Overnight grown primary culture with optical density 0.6 was taken for further inoculation in Nutrient broth and minimal salt liquid media having pendimethalin (sole carbon and energy source). Growth of bacteria was measured by taking absorbance after 2 h interval till 60 h at 660 nm wavelength.
Extraction of pigment from bacterial isolate
The pigment producing bacterial isolate was further grown in nutrient broth in rotary shaker incubated at 28±2°C for three days. After that, the bacterial cells were harvested by centrifugation at 10000 rpm for 15 min. The pellet was washed with sterile water and spin for 15 min at 8000 rpm. The pellet was suspended with 5 ml of methanol. It was incubated in water bath at 55°C for 15 min until all visible pigments were extracted and centrifuged (8000 rpm) for 15 min. The coloured supernatant was separated and filtered through Whatman filter paper (No.1). The coloured extract was analysed by scanning the absorbance in the wavelength ranging from 300 to 800 nm using the spectrophotometer (Schimadzu UV-Vis 1800).
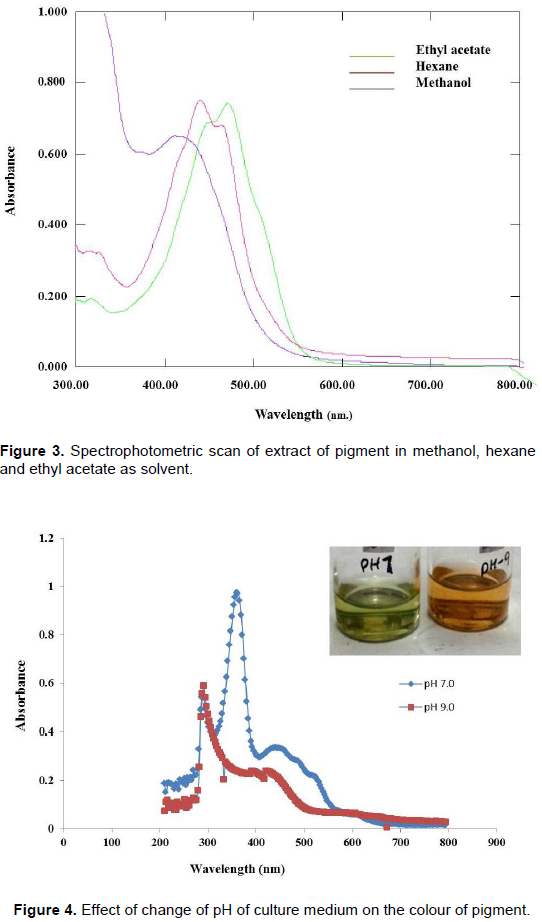
Extraction of pigment in different solvents
The pigment was extracted by different solvents. Primary extraction was done in methanol followed by secondary extraction in hexane and final extraction was done in ethyl acetate in which complete extraction of pigment takes place. Red pigment was extracted in ethyl acetate and yellow pigment in methanol and hexane. Extracted pigments in different solvents were scanned spectrophotometrically to analyse maximum absorbance (λmax). Ethyl acetate was shown to be better than methanol and hexane as the latter two chemicals are not very efficient in the extraction (Henriques et al., 2007).
Effect of change of pH of growth medium on pigment production
Pigment producing bacterial isolate was allowed to grow in nutrient broth medium at pH range 5, 7, 9, and 11 for 3 days. After incubation, pigment was extracted by the same method as described earlier and extracted pigment was analysed spectrophotometrically.
Fourier transform infrared spectroscopy (FTIR) analysis of extracted pigment
The methanolic extract of pigment with optical density (1.5) was taken for FTIR analysis. Beta carotene with a concentration of 1 mg/ml (methanol as solvent) was taken as standard. A quantity of 10 µl of pigment and beta carotene was placed on a diamond window of the spectrophotometer under standard room temperature. A 32 scans with a resolution of 4 cm-1 was adapted. The available spectrum ranges were 400 to 4000 cm-1.
A 16S rRNA based identification and phylogenetic analysis
Total bacterial genomic DNA was isolated using DNA extraction kit (Himedia, India). The amplicon size of 1500 nt fragment of 16S rRNA was amplified using specific primer, forward Gm 3f (5'-AGAGTTTGATCMTGG-3') and reverse Gm 4r (5'-TACCTTGTTACGACTT-3') targeting 16S rDNA. PCR amplified product was purified using DNA purification kit, Himedia, India; HiPuraTM. The purified product was sent for sequencing. The consensus sequences were aligned and analysed for determination of specific bacterium using the NCBI web-based BLAST program (http:// www.ncbi.nlm.nih.gov/BLAST/). The closest known species were compared with nucleotide identity matrix. The nucleotide sequences were aligned using the clustal W program. The phylogenetic analysis of the 16S rDNA sequences of the isolate was conducted with MEGA 5.1 using neighbour- joining method with 1,000 bootstrap replicates (Tamura et al., 2011).
In the present study, a bacterial strain from soil sample was isolated by serial dilution and studied for pigment production. Yassin and Almouqatea (2010) suggested “open plate technique” is suitable for microbial isolation particularly for pigment producing bacteria. The isolation of carotenoid producing microbes from some abnormal environmental condition was also reported by Ramasamy and Udayasuriyan (2006).
Based on coloured colonies production on Nutrient Agar plates, isolate PD5 was selected. The nutrient agar plate shows red colonies with no fluorescence emission under ultra violet (UV) light. It grows at 28 to 40°C, but the optimum temperature is 30°C. It can grow at pH 6 to 10, but optimum growth was found at pH 7. Growth study of bacterial isolate showed that it is able to grow in MSM having pendimethalin as carbon source at 50 mg/L concentration (Figure 1). Identification of bacterial isolate was done using 16S Rrna based polymerase chain reaction (PCR) amplification followed by sequencing.
The sequence data of the 16S rRNA was subjected to BLAST analysis. As 16S rRNA gene sequence provide accurate grouping of organism even at subspecies level, it is considered as a powerful tool for the rapid identification of bacterial species (Jill and Clarridge, 2004). The sequence analysis of 16S rRNA sequences of isolate PD5 showed its maximum identity of 99% to Bacillus subtilis (Figure 2). The 16S rRNA sequences of the bacterial isolate were submitted to NCBI under the accession number (KY306687).
Extract of pigment in different solvents showing characteristic spectra (Figure 3). Methanol extract of the pigment shows λmax at 490 nm, nearer to beta carotene spectra. Change in pH of culture medium has effect on colour. Spectrophotometric scan showed that at pH 7, pigment extract were green in colour at λmax 290 nm, while at pH 9, orange pigment at λmax 310 nm was observed (Figure 4).
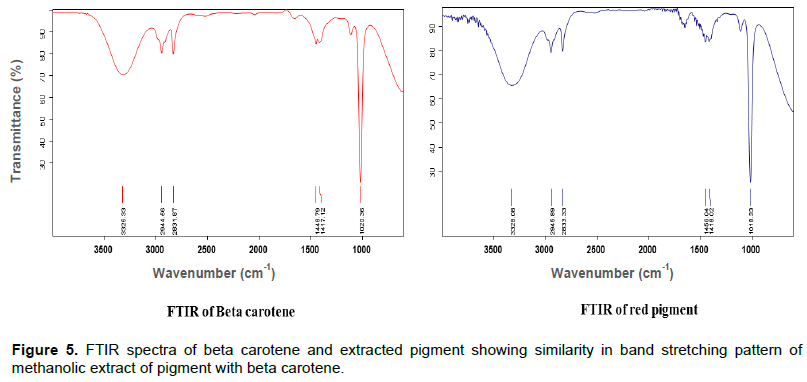
FTIR technique is used to obtain an infrared spectrum, emission, photoconductivity of a solid, liquid or gas. FTIR that operates in the mid infrared region (4000 to 400 cm-1) is a powerful tool for quantitative analysis of fats, oils and palm carotene (Moh et al., 1999). The bands of the characteristic functional groups (CH3, CH2, C=C, C=O, OH, etc.) can be assigned when possible. Some special functional groups such as C=C=C, ‘cross epoxides', etc., which cannot be easily identified by give full form (1H-NMR) methods, can be detected in the FTIR spectra (Lorand et al., 2002).
FTIR spectrum of extracted pigment from bacterial isolate, showed similarity with beta carotene in fingerprinting patterns of peaks (Figure 5). It indicates that extracted pigment is a beta carotene or carotenoid derivative. Analysis of pigment highlighted the stretching of different functional groups with peaks at 1661, 1653, 1654 and 1655 cm-1, respectively. FTIR peaks in the range of 1600 to 1670 correspond to protein (Suresh et al., 2016). The next bands were approximate at 1425, 1426 and 1424 cm-1, usually caused by the bending and vibration of methylene -CH2 (scissoring) seen in beta carotene standard at 1450.68 cm-1 peak height. This could be attributed to lycopene pigments (Parlog, 2011). Bacillus cereus group members are found to be versatile producers of secondary metabolites, such as antimicrobial substances (Abriouel et al., 2011), extra cellular enzymes (Liang et al., 2013) or fluorescent pigments (Banerjee et al., 2013). Melanin producing Bacillus weihenstephanensis isolated from the soil can offer considerable benefit for commercial production of biopigments like carotenoids, anthraquinone, chlorophyll, melanin, flavins, quinones, prodigiosins, monascins, violacein, etc (Drewnowska et al., 2015).
The red pigment producing bacterium isolated from rice field was characterized based on morphology, coloured colony production on Nutrient Agar plate and identified by 16S rRNA gene sequencing. The bacterial strain was identified as a novel strain of Bacillus species. The isolated strain produced intracellular red pigment in nutrient broth medium. The optimum conditions for pigment production were determined which revealed the maximum production of pigment at 30 to 37°C and at pH 7 after 3 days. The red color was extracted with methanol and the extract was analyzed by scanning the absorbance with a UV-VIS spectrophotometer. FTIR analysis of pigment showed similarity with chromatogram given by beta carotene. These results suggest that the pigment producing B. subtilis PD5 can be used for the commercial production of pigment. This strain is able to grow in minimal salt media, so pigment production by fermentation can be commercialized at low cost.
The authors have not declared any conflict of interests.
REFERENCES
|
Abriouel H, Franz CMAP, Omar NB, Gálvez A (2011). Diversity and applications of Bacillus bacteriocins. FEMS Microbiol. Rev. 35:201-232.
Crossref
|
|
|
|
Banerjee D, Mondal A, Gupta M, Guhna AK, Ray L (2013). Optimization of fermentation conditions for green pigment production from Bacillus cereus M116 (MTCC5521) and its pharmacological application. Lett. Appl. Microbiol. 58:25-30.
Crossref
|
|
|
|
|
Drewnowska JM, Zambrzycka M, Kalska-Szostko B, Fiedoruk K, Swiecicka I (2015). Melanin-like pigment synthesis by soil Bacillus weihenstephanensis isolates from North-eastern Poland. PloS One 10(4):e0125428.
Crossref
|
|
|
|
|
Dufossé L (2009). Microbial and Microalgal Carotenoids as Colourants and supplements. Carotenoids 5:83-98.
Crossref
|
|
|
|
|
Fabre CE, Santerre AL, Loret MO, Baberian R, Pareilleux A, Goma G, Blanc PJ (1993). Production and Food Applications of the Red Pigments of Monascus ruber. J. Food Sci. 58(5):1099-1102.
Crossref
|
|
|
|
|
Henriques M, Silva A, Rocha J (2007). Extraction and quantification of pigments from a marine microalga: a simple and reproducible method. Communicating Current Research and Educational Topics and Trends in Applied Microbiology Formatex, 586-593.
View
|
|
|
|
|
Liang TW, Liu CP, Wu C, Wang SL (2013). Applied development of crude enzyme from Bacillus cereus in pre-biotics and microbial community changes in soil. Carbohydr. Polym. 92(2):2141-2148.
Crossref
|
|
|
|
|
Lorand T, Deli J, Molnar P, Toth G (2002). FT-IR study of some carotenoids. Helv. Chim. Acta 85(6):1691-1697.
Crossref
|
|
|
|
|
Mekhael R, Yousif SY (2009). The role of red pigment produced by Serratia marcescens as antibacterial and plasmid curing agent. J Duhok Univ. 12(1): 268-274.
|
|
|
|
|
Moh MH, Che Man Y .B, van de Voort FR, Abdullah WJW (1999). Determination of peroxide value in thermally oxidized crude palm oil by near infrared spectroscopy. J. Food Lipids 76(1):19-23.
Crossref
|
|
|
|
|
Parlog RM (2011). Metabolomic studies applied on different sea buckthorn (Hippophae Rhamnoides L.) varieties, Ph.D. Thesis Wageningen University at Netherlands.
View
|
|
|
|
|
Pattnaik P, Roy U, Jain P (1997). Biocolours: new generation additives for food. Indian Food Ind. 16(5):21-25.
|
|
|
|
|
Ramasamy AK, Udayasuriyan V (2006). Forming Bacterium for Carotenogenesis. Biotechnology 5(1):79-82.
|
|
|
|
|
Sambrook J, Fritsch EF, Maniatis J (1989). Molecular cloning. In: A Laboratory Manual (2nd ed.). USA: Cold Spring Harbor Laboratory.
|
|
|
|
|
Suresh S, Karthikeyan S, Jayamoorthy K (2016). FTIR and multivariate analysis to study the effect of bulk and nano copper oxide on peanut plant leaves. J. Sci. Adv. Mater. Devices 1(3):343-350.
Crossref
|
|
|
|
|
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol 28:2731-2739.
Crossref
|
|
|
|
|
Yassin MF, Almouqatea S (2010). Assessment of airborne bacteria and fungi in an indoor and outdoor environment. Int. J. Environ. Sci. Technol. 7(3):535-544.
Crossref
|
|