ABSTRACT
Cultivation and commercial utilization of genetically modified (GM) crops has significantly increased in many parts of the world and particularly in developing countries where food security is a challenge. Despite stringent regulations requiring that food made from GM foods should be properly labelled, evidence of unlabelled foods made from GM crops sold in local markets in many of these countries is increasing. This challenge provides the justification for the development of a reliable, accurate and effective screening method. Polymerase chain reaction (PCR)-based method for detection of specific genes in GM crops is a common method used in many parts of the world. This study adapted a PCR-based technique to screen for the presence of specific DNA markers of genetic modification in finished maize and soya products collected from selected supermarkets and local markets across Nigeria. Results obtained indicated that 26.7% of samples tested contain GM specific genes. GM specific genes were also detected in some made-in-Nigeria processed food samples. The findings indicate that products made with GMO materials have entered the food chain in Nigeria at a modest scale and identifies the need for capacity building in techniques for GMO detection for regulatory agencies in Nigeria.
Key words: Genetically modified (GM) organism, GM maize, GM soy, polymerase chain reaction (PCR), CaMV35S, Nigeria.
Over the last decade, concerns about food and environmental safety have increased considerably due to the introduction of techniques for genetic modification of organisms into agriculture and food production (Boldura and Popescu, 2016). While an increasing body of scientific evidence indicates reduction of hunger and starvation as well as resistance to pests and drought as part of the beneficial effects of genetic modification of food crops, concerns about health and environmental implications of generation of genetically modified organisms (GMOs) abound. Sateesh (2008) reported that consumption of GMOs could lead to known and unknown risks to human health and the environment, including introduction of allergens and toxins to human beings, transfer of antibiotic resistance markers or unintended transgenes through cross pollination. Due to these concerns, the European Commission (2003a, b) advocates the labeling of GMO products to ensure effective monitoring and traceability. However, this requires accurate and efficient methods for detecting foreign genetic materials or other forms of genetic modification in raw materials as well as in highly processed food.
In many countries, biosafety assessment of GMOs or products made from them is required to assess the potential environmental and health impacts on consumers. This risk assessment demonstrates if unauthorized and potentially unsafe GM products are found in markets (Okpara et al., 2016). GM herbicide-tolerant and insect-resistant soybeans, maize, cotton, and rape seeds are now commonly grown in many parts of the globe and this has further contributed to the global nature of the health and safety concerns of products made from GM crops (SönmezoÄŸlu and Keskin, 2015).
The detection of foreign DNA or identification of changes in DNA composition has been recognized as the most effective tool for GM food examination. This is because the DNA is the most stable molecule during food processing (Datukishvili et al., 2015). Amplification of the promoters, terminators, or inserted transgenes by polymerase chain reaction (PCR) using specific primer sets is the gold standard method for GMO detection (Broeders et al., 2012; Milavec et al., 2014). GM soybean and maize are the most widely distributed transgenic crops worldwide. Therefore, several PCR-based methods for detecting genetic modification in these crops are available in literature. Kim et al. (2013) reported qualitative multiplex PCR methods for identifying different lines of GM maize or GM soybean using event-specific primers. Matsuoka et al. (2000) also described methods which target several frequently used foreign DNA segments (including transgenes, promoter, and terminator) in a single-locus PCR-based method.
The use of three GMO-specific primer pairs directed toward the cp4-epsps transgene, 35S promoter and NOS terminator together with two soybean-specific primer pairs targeting lectin and b-actin genes has been reported by James et al. (2003). Similarly, Forte et al. (2005) developed a molecular screening method based on multiplex-PCR that involves amplification of the 35S promoter as well as the NOS terminator for the detection of GM soya and maize.
Concerns associated with increasing availability of GM plant products in many developing countries underscores the need for the development of appropriate techniques for GMO detection, which allows proper identification, monitoring and enforcement of extant regulations on labelling GM products. In a country such as Nigeria, such validated technique will become a useful tool for appropriate government agencies and help in the making of informed biosafety decisions. This study attempted the adaptation of the PCR-based technique, which uses DNA markers targeting important GMO specific sequences such as cauliflower mosaic virus CaMV 35S promoter, NOS terminator, 5-enolpyruvylshikimate-3-phosphate synthase (EPSPS) gene for herbicide tolerance in soybeans and Bt resistance gene (cry1Ab) in maize, to screen for the presence of GM material in finished maize and soya products in selected supermarkets and local markets across Nigeria.
Sample collection
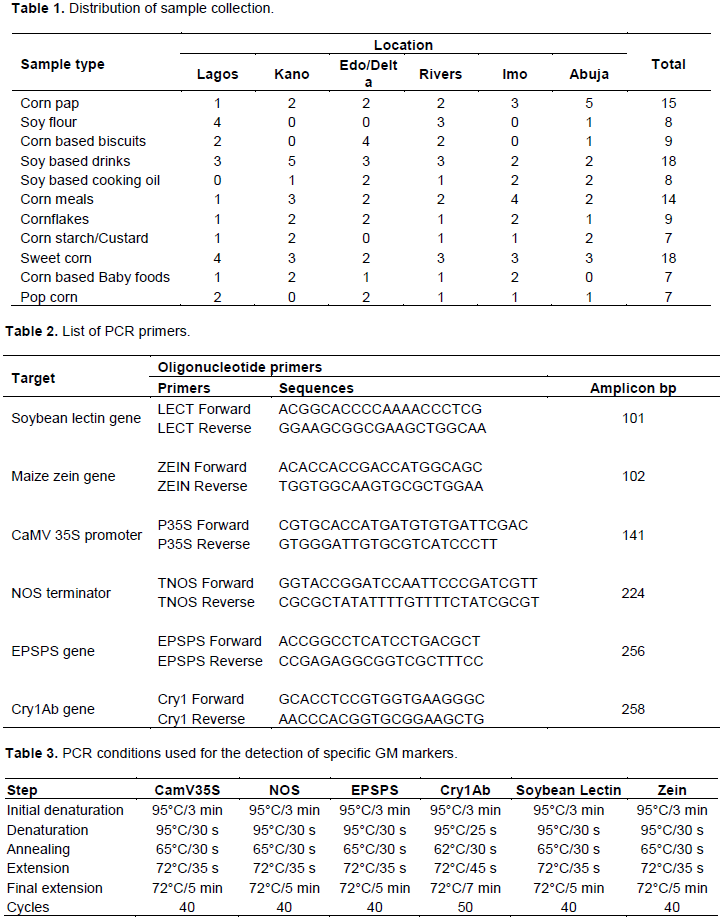
Finished or semi-finished imported food products, made from corn or soybean, were randomly collected across markets in the six geo-political zones of Nigeria. These include products such as corn flour, soybean flour, biscuits, drinks, cooking oil, whole meals, cornflakes, cracker, corn chips, corn starch, popcorn, sweet corn, baby foods, and soy cakes. The sampling strategy targeted products which are in use by the general public. A total of 120 samples were collected in this study. Product sampling was carried out in 12 local markets and 12 major super markets across Nigeria (Table 1). All products were properly labelled for easy identification.
DNA extraction and purification
Genomic DNA extraction was carried out using the CTAB method described by Doyle and Doyle (1987) with slight modifications. Briefly, the protocol used 700 μl CTAB buffer for initial incubation, 500 μl isopropanol for DNA precipitation and 100 μl 70% ethanol for the two washing steps. Finally, extracted DNA samples were dissolved in 200 μl TE buffer supplemented with 10 mg/ml RNase (2 μl). Where necessary, DNA extraction was carried out using the commercially available Zymo Research Plant and Seed Extraction Kits following the manufacturer’s recommended procedure. Purity and quantification of DNA samples were assessed using a UV spectrophotometer. The DNA samples were stored at -20°C until used for PCR.
Primer sequences
Four GMO specific DNA markers sequences, such as cauliflower mosaic virus (CaMV) 35S promoter, NOS terminator, herbicide tolerant EPSPS gene in soybean and insect control GM trait (Cry1Ab gene) were targeted in this study in line with previous studies (Datukishvili et al., 2015). Soy-specific lectin gene and maize specific zein gene were also amplified in relevant samples. Details of GMO specific sequences amplified and the list of primers used in this study are summarized in Table 2. Primers were purchased in lyophilized form from Eurofins MWG Operon (Germany) and reconstituted as directed by the manufacturer.
PCR detection of marker sequences
The PCR reaction mix (20 μl) contained 12 μl OneTaq Quick Load PCR Master Mix (New England Biolabs; 20 mM TrisHCl, 1.8 mM MgCl2, 200 μM dNTPs), 1 μl Taq DNA polymerase (25 units/ml), 1 μl bovine serum albumin (10 mg/ml), 5 μl template DNA and 1 μl each of forward and reverse primers. Reactions were carried out in triplicates in order to confirm amplifications. PCR reactions were performed using a PTC 0200 DNA Engine (Bio-Rad, USA) themocycler. For all genes except Cry1Ab, the PCR cycling condition involves an initial denaturation of 95°C for 3 min, followed by 40 cycles of denaturation at 95°C for 30 s, annealing at 65°C for 30 s, elongation at 72°C for 35 s and a final extension step at 72°C for 5 min (Table 3). The PCR cycling conditions for Cry1Ab is slightly different and consists of initial denaturation at 95°C for 3 min, followed by 50 cycles of denaturation 95°C for 25 s, annealing at 62°C for 30 s, elongation at 72°C for 45 s and a final extension step at 72°C for 7 min.
Agarose gel electrophoresis
PCR products were checked for amplification of target genes using agarose gel (1%) electrophoresis at 140 V for 30 min. DNA bands were captured using the gel documentation system (Edvotek, USA). All results were analyzed visually on the gel documentation system.
This study screened for the presence of DNA markers of specific GMO sequences in 120 samples made from maize or soybeans collected from different locations in Nigeria. Due to the immensely important implications of results obtained in investigations such as this, appropriate quality control to avoid false positive or false negative results is vital (Hübner et al., 1999; Arun et al., 2013). In this study, false positive results were eliminated at two levels. In the first instance, sterile water was processed in parallel to all samples at each step of the extraction protocol in order to eliminate false positive results that may arise due to contamination during DNA extraction. Secondly, no template controls were run to eliminate contamination during the PCR amplification step of this protocol. To ensure that samples showing negative results for CaMV 35S and NOS genes contain sufficient DNA, soy-specific lectin and maize-specific were also analysed. These genes are present in both GM or non-GM soybean and maize, respectively (Zimmermann et al., 1998; Nguyen et al., 2009; Gabriadze et al., 2014).
DNA extraction
Successful screening for the presence of GMO sequences in samples, such as those used in this study, depends largely on the robustness of the DNA extraction method (Tengel et al., 2001; Ahmed, 2002). Sufficient DNA quantity was extracted from all samples used in this study except soy-based cooking oil and corn-based baby food (Table 4). DNA yield obtained ranged between 2.2±0.17 ng/µl obtained for sweet corn and 23.8±0.22 ng/µl obtained for popcorn. The A260/280 ratio obtained for all the samples ranged between 1.64 and 1.88, which indicated that they are of acceptable quality. This result is consistent with reports of previous studies which extracted DNA from processed food products (Lipp et al., 1999; Cardarelli et al., 2005; Greiner and Konietzny, 2008; Mandaci et al., 2014). According to Jasbeer et al. (2008), CTAB DNA extraction method is efficient and widely used for the extraction of pure DNA plants and plant derived food products due to its ability to provide effectively separate plant DNA from polysaccharides. However, DNA yield obtained in this study is lower as compared to values reported for similar studies (Mandaci et al., 2014). It is not yet clear if the poor yield observed results from the modification of the CTAB protocol carried out in this study or due to important food processing factors, such as temperature and pH, which affect DNA quality and quantity in processed food materials (Gryson, 2010).
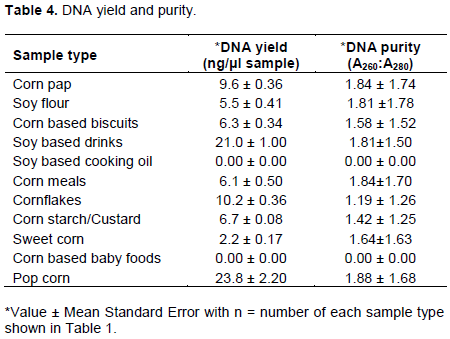
The inability to extract DNA from processed soybean oil is consistent with the available (Costa et al., 2009). Moreover, it is also possible that the DNA in the corn- based baby food products has been destroyed during processing food-processing conditions such as temperature and pH could lead to DNA degradation which makes PCR analysis of the DNA thus rendering PCR analysis impossible (Ahmed, 2002; Gryson, 2010; Okpara et al., 2016). DNA materials extracted in this study are also visible on agarose gel (Figure 1).
PCR amplifications
In this study, zein gene was successfully amplified from the DNA samples obtained from maize-based products and a similar trend was observed for the amplification of lectin gene DNA samples isolated from soy-based products (Table 5). Agarose gel electrophoretic analysis of these samples also produced visible bands for zein and lectin gene in respective samples (Figures 2 and 3). These results indicate that isolated DNAs are amplifiable and contained maize or soy DNA, respectively. The result also validates data obtained for the PCR amplification of the GMO specific genes targeted despite the low DNA yield obtained in this study.
Positive results for the detection of GMO specific genes were observed for 28 samples, representing 26.7% of all samples screened in this study (Figure 3). This is made up of 19 maize- and 9 soy-based samples. This result indicates that 24% of all maize-based products and 34% of all soy-based samples showed positive results for the presence of GMO specific genes. The present study showed consistency in the detection of both CaMV35S and NOS terminator genes for all GMO-positive samples. The percentage of maize samples harbouring CaMV35S and NOS sequences observed in this study (24%) is lower when compared with 28% previously reported by Okpara et al. (2016). This discrepancy may be due to the fact that Okpara et al. (2016) screened 61 samples collected only from the southern part of Nigeria as compared to 120 samples collected from the six regions of Nigeria. The southern part of Nigeria hosts all the major seaports and represents the entry point for all important food products in Nigeria. This coupled with infrastructural challenges facing food transportation within the country may partly account for the higher percentage of GMO-containing food products in the southern part of Nigeria, as observed by Okpara et al. (2016). No previous studies have screened soy-based products for the presence of CaMV35S and NOS sequences and the present result represents a baseline data for future investigations.
Genetic modification in transgenic maize (corn), focused on the production of crops with agronomic desirable traits, include the incorporation of a gene that codes for the Bacillus thuringiensis toxin (Cry1ab gene) in the plant’s genome. Plants harbouring this genetic modification express the Cry1ab protein and are resistant to insect pests. In addition, herbicide (glyphosate and glufosinate)-tolerant plants have also been produced (OECD, 2002). In this study, roundup ready soybeans, which expresses the 5-enolpyruvylshikimate-3-phosphate synthase (EPSPS) gene, is one of the first series of GM glyphosate-resistant soybeans produced and cultivated globally (Clarke et al., 2013). Screening conducted in this study showed that 35% of soy-based products harbor the EPSPS gene (Figure 4A). Interestingly, only soy-based drink samples tested positive for the presence of CaMV35S, NOS and the EPSPS genes. Cry1ab gene was also successfully amplified in all CaMV35S- and NOS-positive DNA samples screened (Figure 4B). This observation is consistent with the previous report by Okpara et al. (2016) which identified five products made with GM maize. GM cowpea is one of the most widely cultivated GM crop worldwide.
Consistent with other national GMO-screening studies, this study indicate that positive results or GMO-specific genes were observed mostly in products imported into Nigeria (Arun et al., 2013; Mandaci et al., 2014).
However, this study also identified GMO specific genes in some processed foods produced in Nigeria. Maize and soybean are widely cultivated in Nigeria and Nigeria has been reported to be the largest producer of soybean in sub-Saharan Africa (Dugje et al., 2009). Therefore, it was presupposed that raw materials for these made in Nigeria processed foods were sourced locally in Nigeria. This implies that it is possible that GM maize and soybean are also currently being cultivated in Nigeria.
This study has successfully used PCR-based techniques to screen for the presence of GM foods in the Nigerian market. PCR-based techniques for GMO detection is a highly sensitive and globally validated approach for GMO screening (Ahmed, 2002; Anklam et al., 2002; Forte et al., 2005; Meriç et al., 2014). Moreover, GMO-specific DNA markers used in this study are also the universally accepted markers for this type of screening. Therefore, based on the findings of this study, it could be concluded that products made with GMO materials have entered the food chain in Nigeria at a modest scale.
Due to ethical and biosafety concerns, GM foods are strongly regulated in many countries. Theoretically, the consumption of foods containing genetically engineered DNA is expected to be digested without any adverse consequences and there is currently no report of adverse reactions resulting from the consumption of food made from GM crops. Even though public concern of the unknown health risk of GMOs to human and animals abound (Lisha et al., 2017), proponents of GMO foods continue to highlight benefits, including the ability of the technology to tackle the problem of food scarcity in many developing countries. However, the need to have proper labelling through appropriate identification techniques to protect consumers’ right and to ensure food safety has been suggested (Azadi and Ho, 2010). It is also important that regulatory bodies, particularly in countries such as Nigeria, have the capacity to check materials imported for food manufacturing purposes for the presence of GMO specific sequences. In 2015, the Nigerian Government established the National Biosafety Management Agency (NBMA) via the enactment of the National Biosafety Management Agency Act 2015. The agency provides regulatory framework for the introduction of GMOs into Nigeria, recently approved the commercial release of GM cotton and gave permission for confined field trial of GM maize. However, findings of this study together with previous reports by Okpara et al. (2016) presuppose that products made from GM crops have been in the Nigerian market before, prior to the establishment of the NBMA.
The authors have not declared any conflict of interests.
REFERENCES
|
Ahmed FE (2002). Detection of genetically modified organisms in foods. Trends Biotechnology 20:215-223.
Crossref
|
|
|
|
Anklam E, Gadani F, Heinze P, Pijnenburg H, VandenEede G (2002). Analytical methods for detection and determination of genetically modified organisms (GMO's) in agricultural crops and plant-derived food products. European Food Research Technology 21:3-26.
Crossref
|
|
|
|
|
Arun OO, Yilmaz F, MuratoÄŸlu K (2013). PCR detection of genetically modified maize and soy in mildly and highly processed foods. Food Control 32:525-531.
Crossref
|
|
|
|
|
Azadi H, Ho P (2010). Genetically modified and organic crops in developing countries: A review of options for food security. Biotechnology Advances 28:160-168.
Crossref
|
|
|
|
|
Boldura OM, Popescu S (2016). PCR: A Powerful Method in Food Safety Field. In: Samadikuchaksaraei A (Ed.) Biochemistry, Genetics and Molecular Biology "Polymerase Chain Reaction for Biomedical Applications". Intech Publishers, USA. pp. 135-158.
Crossref
|
|
|
|
|
Broeders SR, De-Keersmaecker SC, Roosens NH (2012). How to deal with the upcoming challenges in GMO detection in food and feed. Journal of Biomedicine and Biotechnology 2012:402-418.
Crossref
|
|
|
|
|
Cardarelli P, Branquinho MR, Ferreira RTB, Da Cruz FP, Gemal AL (2005). Detection of GMO in food products in Brazil: the INCQS experience. Food Control 16:859-866.
Crossref
|
|
|
|
|
Clarke JD, Alexander DC, Ward DP, Ryals JA, Mitchell MW, Wulff JE, Guo L (2013). Assessment of genetically modified soybean in relation to natural variation in the soybean seed metabolome. Scientific Reports 3:3082-3087.
Crossref
|
|
|
|
|
Costa J, Mafra I, Amaral JS, Oliveira MBP (2010). Detection of genetically modified soybean DNA in refined vegetable oils. European Food Research and Technology 230:915-923
Crossref
|
|
|
|
|
Datukishvili N, Kutateladze T, Gabriadze I, Bitskinashvili K, Vishnepolsky B (2015). New multiplex PCR methods for rapid screening of genetically modified organisms in foods. Frontiers in Microbiology 6:757-761.
Crossref
|
|
|
|
|
Doyle JJ, Doyle JL (1987). A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Photochemistry Bulletin 19:11-15.
|
|
|
|
|
Dugje IY, Omoigui LO, Ekeleme F, Bandyopadhyay R, Kumar PL, Kamara AY (2009). Farmers' guide to soybean production in northern Nigeria. IITA.
|
|
|
|
|
European Commission (2003a). Regulation (EC) No.1829/2003 of the European Parliament and of the Council of 22 September 2003 on genetically modified food and feed. Official Journal of the European Union 268:1-23.
|
|
|
|
|
European Commission (2003b). Regulation (EC) No.1830/2003 of the European Parliament and of the Council of September 2003 concerning the trace ability and labeling of genetically modified organisms and the trace ability of food and feed products produced from genetically modified organisms and amending Directive2001/18/EC. Official Journal of the European Union 268:24-28.
|
|
|
|
|
Forte VT, Di Pinto A, Martino C, Tantillo GM, Grasso, G, Schena FPA (2005). General multiplex-PCR assay for the general detection of genetically modified soya and maize. Food Control 16:535-539
Crossref
|
|
|
|
|
Gabriadze I, Kutateladze T, Vishnepolsky B, Karseladze M, Datukishvili N (2014). Application of PCR-based methods for rapid detection of corn ingredients in processed foods. International Journal of Food Sciences and Nutrition 3:199-202.
|
|
|
|
|
Greiner R, Konietzny U (2008). Presence of genetically modified maize and soy in food products sold commercially in Brazil from 2000 to 2005. Food Control 19:499-505.
Crossref
|
|
|
|
|
Gryson N (2010). Effect of food processing on plant DNA degradation and PCR-based GMO analysis: a review. Analytical and Bioanalytical Chemistry 396:2003-2022.
Crossref
|
|
|
|
|
Hübner P, Studer E, Häfliger D, Stadler M, Wolf C, Looser M (1999). Detection of genetically modified organisms in food: critical points for quality assurance. Accreditation and Quality Assurance 4:292-298.
Crossref
|
|
|
|
|
Jasbeer K, Ghazali FM, Cheah YK, Son R (2008). Application of DNA and Immunoassay Analytical Methods for GMO Testing in Agricultural Crops and Plant-Derived Products. ASEAN Food Journal 15:1-25
|
|
|
|
|
James D, Schmidt AM, Wall E, Green M, Masri S (2003). Reliable detection and identification of genetically modified maize, soybean and canola by multiplex PCR analysis. Journal of Agricultural and Food Chemistry 51:5839-5834.
Crossref
|
|
|
|
|
Kim JH, Jeong D, Kim YR, Kwon YK, Rhee GS, Zhang D (2013). Development of a multiplex PCR method for testing six GM soybean events. Food Control 31:366-371.
Crossref
|
|
|
|
|
Lipp M, Anklam E, Brodmann P, Pietsch K, Pauwels J (1999). Results of an interlaboratory assessment of a screening method of genetically modified organisms in soy beans and maize. Food Control 10: 379-383.
Crossref
|
|
|
|
|
Lisha V, New CY, Nishibuchi M, Son R (2017). Rapid genetically modified organism (GMO) screening of various food products and animal feeds using multiplex polymerase chain reaction (PCR). Journal of Food Research 1:1-8.
Crossref
|
|
|
|
|
Mandaci M, Çakir Ö, Turgut-Kara N, Meriç S, Ari, Åž (2014). Detection of genetically modified organisms in soy products sold in Turkish market. Food Science and Technology 34:717-722.
Crossref
|
|
|
|
|
Matsuoka T, Kawashima Y, Akiyama H, Miura H, Goda Y, Kusakabe Y., Isshiki K, Toyoda M, Hino A. (2002). Detection of recombinant DNA segments introduced to genetically modified maize (Zea mays). Journal of Agricultural and Food Chemistry 50:2100-2109.
Crossref
|
|
|
|
|
Meriç S, Çakır Ö, Turgut KN, Arı Åž (2014). Detection of genetically modified maize and soybean in feed samples. Genetics and Molecular Research 13:1160-1168.
Crossref
|
|
|
|
|
Milavec M, Dobnik D, Yang L, Zhang D, Gruden K, Zel J (2014). GMO quantification: valuable experience and insights for the future. Analytical and Bioanalytical Chemistry 406:6485-6497.
Crossref
|
|
|
|
|
Nguyen CT, Son R, Raha AR, Lai OM, Clemente-Michael WVL (2009). Comparison of DNA extraction efficiencies using various methods for the detection of genetically modified organisms (GMOs). Food Research International 16:21-30.
|
|
|
|
|
Organization for Economic Co-operation and Development (OECD), (2002). Consensus document on compositional considerations for new varieties of maize (Zea mays): Key food and feed nutrients, anti-nutrients and secondary plant metabolites. ENV/JM/MONO (2002) 25. Series on the Safety of Novel Foods and Feeds, No. 6. Paris, France. 2002. Available at:
View
|
|
|
|
|
Okpara CN, Elijah AI, Adamu LOG, Uzochukwu SVA (2016). Screening for genetically modified maize in raw and processed foods sold commercially in Southern Nigeria Border States. Applied Food Biotechnology 3:50-158.
|
|
|
|
|
Sateesh MK (2008). Use of genetically modified organisms and their release in the environment. In: Sateesh, MK (Ed.) Bioethics and Biosafety. New Delhi: I.K. International Pvt. Ltd. pp. 217-242.
|
|
|
|
|
SönmezoÄŸlu OA, Keskin H (2015). Determination of genetically modified corn and soy in processed food products. Journal of Applied Biology and Biotechnology 3:32-37.
|
|
|
|
|
Tengel C, Schübler P, Setzke E, Balles J, Sprenger-Haubles M (2001). PCR based detection of genetically modified soybean in maize in raw and highly processed foodstuffs. BioTechniques 31(2):426-429.
Crossref
|
|
|
|
|
Zimmermann A, Luthy J, Pauli U (1998). Quantitative and qualitative evaluation of nine different extraction methods for nucleic acids on soya bean food samples. Zeitschrift Für Lebensmittel-Untersuchung Und-Förschung 207:81-90.
Crossref
|
|