ABSTRACT
Lipases hydrolyze lipids to yield fatty acids and glycerol which are used as raw materials in industries. The lipase from raphia mesocarp was extracted, purified and characterized. Assay was carried out using p- nitrophenyl palmitate as substrate. The lipase was subjected to 80% ammonium sulphate precipitation, purified on ion exchange and gel filtration chromatography. Native and subunit molecular weights were determined on a gel filtration column and 10% sodium dodecyl sulphate polyacrylamide gel electrophoresis respectively. Kinetic parameters (Km and Vmax), effect of temperature, pH, sensitivity to metal ions and substrate specificities of the lipase enzyme were studied. Specific activity of 17.40µmol/min/mg and purification fold of 2.68% were obtained.Native and subunitmolecular weightwas 35 kDa. The Km and Vmax values were0.01mM and 20.5µmol/min/ml respectively. The lipase enzyme was inhibited by all metallic salts used and enhanced by CaCl2. Optimal pH was 7.0 and temperature for maximal activity was 40ï‚°C. The lipase hydrolyzed coconut oil at about twice the rate of palm kernel oil and palm oil, and at a higher rate than olive oil and Raphia oil. The study reveals that raphia mesocarp isa possible source of lipase which can be of industrial use.
Key words: Lipase, raphia palm fruit, mesocarp
Lipases are a group of enzymes that catalyses the hydrolysis (cleavage) and synthesis of various forms of lipids (Svendsen, 2000). They belong to a subclass of the esterases and have been reported to be active at the oil water interface (Ibrahim, 1996). Oilseed lipases have great potential for commercial exploitation as industrial enzymes. The demand of lipolytic enzymes has been increased due to its potential use in the various manufacturing processes of industrial goods such as food and pharmaceuticals (Boland et al., 1991; Gandhi, 1997), in the production of biodiesel and industrial detergent (Freire and Castilho, 2008) as well as in the analysis of triacylglycerols (Forgia et al., 1995), raw materials in food, cosmetics and pharmaceutical industries (Hermansyah et al., 2006). Fat splitting, with lipase as a catalyst, is advantageous compared to conventional process due to low energy consumption, low cost, high product quality, ease of purification and safer process. The yield of plant lipases is low; sometimes it could be cheaper than other sources and does not require fermentation.
Lipase activity has been reported in several oil plant extracts which include castor bean, palm seeds, oleifera seed, sunflower seed, peanuts (Abigor et al., 2002; Henderson and Osborne, 1991; Khan et al., 1991) and palm fruit mesocarp (Abigor et al., 1985).
Raphia is an underutilized and under exploited plant that produces fruits, with improved agronomical practices; tons of the fruits can be produced for cheap source of lipase which can be used industrially. In this study we report the isolation, purification and characterization of lipase from raphia palm mesocarp to hydrolyze vegetable oils for production of fatty acid and glycerol.
Raphia palm fruits were obtained from Mile 18 village, Edo State, Nigeria. Palm oil, coconut oil, palm kernel oil used for the hydrolysis reaction were obtained from Mega stone farms, Oba town, Ogun State, Nigeria. Raphia oil was extracted by soxhlet using n-Hexane as solvent and olive oil was bought from a supermarket in Benin, Edo State, Nigeria. All chemicals, reagents, resins, standard protein and solvents used in the experiment were of analytical grades and were obtained from Sigma chemical Company, St Louis, Mo., USA, Pharmacia Fine Chemical, Uppsala, Swedenand BDH Chemicals Limited, Poole, England.
Enzyme preparations
The Raphia fruit was allowed to ripe, the exocarp was peeled off and the mesocarp was scraped with a cleaned knife and homogenized for 5 min in 0.15M Tris buffer adjusted to pH7.5 with KOH. The homogenate was filtered through one layer of cheese cloth and centrifuged for 30min at 10,000g using High Speed Refrigerated Centrifuge. It yielded a fat layer, a supernatant liquid and a pellet. The supernatant was used for the assay of lipase activity and protein concentration determination. The remaining supernatant was fractionally precipitated using ammonium sulphate.
Assay of enzyme activity
Lipase activity was determined by the colorimetric method according to Lotrakul and Dharmsthiti (1997). This method was based on the cleavage of p-nitrophenyl palmitate (p-NPP) at pH 8.0. The reaction contained 180µl of solution A (0.062g of p-NPP in 10ml of 2-propanol, sonicated for 2 min before use), 1620µl of solution B(0.4% triton X-100 and 0.1% gum Arabic in 50 mM Tris-Hcl, pH 8.0) and 200µl of properly diluted enzyme sample. The product was detected at 410 nm wavelength after incubation for 15min at 37°C. Under this condition, one unit of lipase activity (U) was defined as the amount of enzyme required to release 1µmol of p- nitrophenol (p-NP) per minute under standard assay condition.
Protein determination
Protein concentrations were determined according to Lowry’s method (1951) using BSA (1mg/ml) as the standard protein, absorbance was read at 660 nm and a standard graph was plotted. The amount of protein present in the sample was estimated from the standard graph.
Enzyme purification
The crude extract from the centrifugation step was fractionally precipitated to 80% ammonium sulphate saturation (516 g/L) according to the method of Doonan (1996). After standing for 1 h in an ice bath, the suspension was centrifuged at 10,000g for 15 min at 4°C, the supernatant discarded, and the precipitate collected and was dialyzed against Tris buffer for 4 h to remove traces of ammonium sulphate.
Ion exchange chromatography was performed using DEAE Sephadex A-50 as the resin. The resin was packed into a 1.5 x 40 cm column. 5ml of the dialyzed extract was carefully layered on the column bed surface that has been previously equilibrated with about 3 L of Tris buffer, pH 7.5, containing 1mM EDTA and was allowed to drain into the bed. The column was washed with Tris buffer to remove unbound protein, followed by elution with stepwise gradient of 1M NaCl in Tris buffer. Elution was performed at a flow rate of 30ml/h and fraction of 5ml each was collected. Protein was monitored spectrophotometrically at 280nm. The fractions were also assayed for lipase activity. The fractions having lipase activity were pooled stored in the refrigerator for further purification.
The enzyme fraction was further purified on gel filtration chromatography using Sephadex G-200 as the resin. 5ml of the DEAE-Sephadex sample was layered on the column (1.5 × 40cm). Elution was performed at a flow rate of 10ml/h and fractions of 5ml each were collected. Protein was monitored spectrophotometrically at 280nm. The fractions were also assayed for lipase activity. The fractions having lipase activity were pooled.
Determination of native molecular weight
The native molecular weight of lipase wasdetermined by gel filtration on a column of Sephadex G-100 (1.5 x 100 cm) using the followingproteinmarkers:BlueDextran(2,000Da),GammaGlobulin(150,000Da), Creatine phosphokinase (81,000Da), Bovine serum albumin(68,000Da),Ovalbumin(45,000Da),AlphaChymotryp-sinogen(25,700Da).Void volume (Vo) was determined using blue dextran. Apparent molecular weight was determined by semi logarithmic plot of Ve/Vo against molecular weight of the marker proteins.
Determination of subunit molecular weight
The subunit molecular weight of purified lipase extract was analyzed with sodium dodecyl sulphate polyacrylamide gel electro-phoreses (SDS-PAGE) according to the method of Laemmli (1970) in 12% slab gel. About 0.1ml of purified protein was loaded onto the gel. The gel was stained with 0.1% Coomassie brilliant blue R-250.
Determination of kinetic parameters
The Michelis-Menten constant (Km) and the maximum velocity (Vmax) were determined by measuring enzyme activity of the purified lipase at various concentrations of emulsified substrates. The kinetic parameters (Km and Vmax) were calculated by plotting 1/V against 1/[S] in Lineweaver Burk plot (1934).
Effect of temperature on enzyme activity
A thermostated, water-jacketed reaction chamber was employed in determining the temperature dependence of lipase
activity. The optimum temperature was determined by measuring the lipase activity at different temperatures of 30, 40, 50, 60, and 70°C after 1 h incubation at pH 7.5 at standard conditions. The values of lipase activities were plotted against temperature.
Thermostability of lipase
Thermal stability was determined by incubating the enzyme at different temperatures ranging from 30 to 70°C for various length of time (30, 60, 90, 120, and 150 minutes) at pH 7.5. Lipase activity was measured and expressed as a percentage of the lipase activity.
Effect of pH on enzyme activity
The effect of pH on the activity of purified enzyme was studied by measuring the enzyme activity at various pH range of 3.5 to 10.0 for 1 h at room temperature. The lipase activity was assayed under standard conditions.
Enzyme inhibition
The following salts at the concentration of 5– 20mM were tested for their possible inhibitory effects on the activity of the purified lipase enzyme: zinc chloride, manganese chloride, calcium chloride, potassium chloride and EDTA at room temperature for 1h and the lipase activities were determined.
Effects of substrates (hydrolysis reactions)
Palm oil, raphia oil, oliveoil, palm kernel oil and coconut oil were used as lipase substrates. The degree of lipolysis was determined by the assay method described previously above.
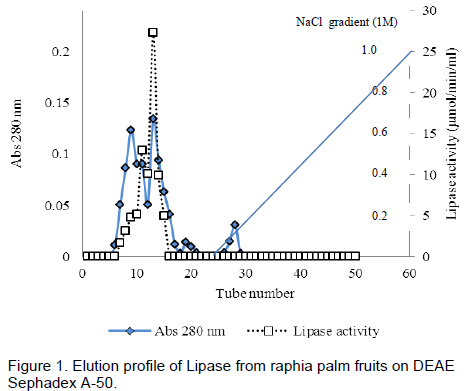
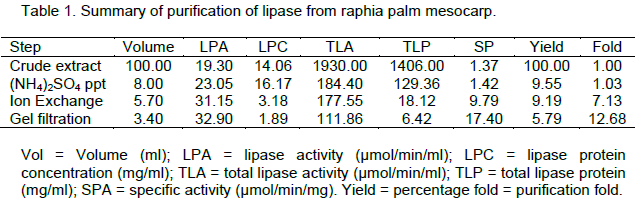
The elution profile of the crude extract from ion – exchange chromatography column of DEAE Sephadex A-50 is presented in Figure 1. The enzyme was not bound to the DEAE-Sephadex A-50 as it came out in the flow through fractions.No lipase activity was recovered in the fractions obtained by gradient elution. The crude protein was adsorbed on the DEAE- Sephadex A-50 column and this was eluted with a stepwise NaCl gradient(Figure 1), single peak was obtained. The active fractions from the peak was pooled together and used for further purification gelfiltrationchromatography. Single peak was also obtained, active fractions were pooled and used to assay the enzyme. A summary of the purification procedures is presented in Table 1. The partially purified enzyme from the mesocarp of raphia had a specific activity of 17.40 µmol/min/mg, a purification fold of 12.68 and a percentage yield of 5.79.
Five millilitre (5ml) of the dialyzed extract was carefully layered on the DEAE Sephadex A-50 column bed (1.5 x 40 cm) surface that has been previously equilibrated with about 3 L of Tris buffer, pH 7.5, containing 1mM EDTA and was allowed to drain into the bed. The column was washed with Tris buffer to remove unbound protein, followed by elution with 1M NaCl in Tris buffer. Elution was performed at a flow rate of 30ml/h and fraction of 5ml each was collected. Protein was monitored spectrophotometrically at 280 nm. The fractions were also assayed for lipase activity. The fractions 7 to 21 were pooled for further purification.
Five millilitre (5 ml) of the DEAE-Sephadex sample was layered on the column bed of Sephadex G-200 (1.5 x 40 cm). Elution was performed at a flow rate of 10ml/h and fractions of 5 ml each were collected. Protein was monitored
spectrophotometrically at 280nm (Figure 2). The fractions were also assayed for lipase activity. The fractions having lipase activity were pooled and used for analysis.
Molecular weights determination
The calibration curve on Sephadex G-100 (1.5 x 100 cm) for the determination of native molecular weight is shown in Figure 3. The nativemolecular weight of lipase from raphia mesocarp was 35 kDa.
The picture and calibration curve for the SDS-polyacrylamide gel electrophoresis on 12% slab gels of lipase from Raphia mesocarp is shown in Figure 4. The molecular weight of lipase was estimated to be 35 kDa.
The Lineweaver Burk plot indicating the (Km and Vmax) of lipase from raphia mesocarp is shown in Figure 5.
The results on effect of temperature, thermostability, effect of pH on lipase from Raphia mesocarp are shown in Figures 6, 7 and 8 respectively. Figure 9 shows the hydrolytic activity of the lipase on some vegetable oils and the effect of salt of metal ions on lipase activity are seen in Figure 10.
The precipitation of the enzyme was carried out using ammonium sulphate since it was highly soluble in water, cheap and had no deleterious effect on structure of protein.
The specific activity obtained from the purification of lipase from Raphia mesocarp after further purification using Sephadex G 200 and DEAE Sephadex A-50 resins was 17.40, yield of 5.79% and purification fold of 12.68. Lipase from Propionibacteriumacidpropionicion Sephadex G-50 has a 33-fold increase in specific activity (Sarada and Joseph, 1992).
The purified lipase was monomeric and had a subunit and native molecular weight of 35 kDa obtained on gel filtration which is similar to the molecular weight of Aspergillus niger lipase 35 kDa on Sephadex G-100 gel filtration chromatography and SDS-PAGE (Sugihara et al., 1988), and also monomeric lipase of Pseudomonasfragi,andPseudomonasfluorescenswithmolecularweights of 33 kDa.
In many cases, lipases appear to obey Michaelis–Menten kinetics (Guit et al.,1991). Michaelis–Menten kinetics are characterized by two parameters, Km and Vmax. The latter is the maximum rate of reaction and Km is a measure of the affinity of an enzyme for a particular substrate. The kinetic parameters (Km and Vmax) of lipase obtained by Lineweaver Burk plot when p-nitrophenyl palmitate was used as substrate were 0.01mM and 20.5µmol/min/mg for the mesocarp of raphia.For a Pseudomonascepacia lipase, Pancreac’h and Baratti (1996) reported Vmax value of30 mmol/min when pNPP was used as substrate. The Km reported in this study is an indication that the enzyme from the germinated palm seeds has high affinity for the substrate and could catalyse the hydrolysis reaction for production of glycerol and fatty acid very fast.
One of the factors that affect the activity of enzymes is temperature and is often used to characterize enzyme. In this study, the optimum temperature of lipase from Raphia mesocarp was 40°C. Activity was rapidly lost at temperatures greater than 50°C. This is similar to the report of Abigoret al.(2003) on lipase from germinating seeds of Jatropha curcas L. Iftikhar et al. (2010c) reported that the optimum temperature for lipase from Rhizopus oligosporus was 40°C. The relative activity of the partial purified cocoa lipase has an optimum temperature range of 30- 40°C (Ratna, 2008).
The thermal stability experiment showed that lipase from this plant source was stable at 40°C for 60 min, retaining about 95% of its activity. The lipase enzyme was observed to be sensitive to thermal activation; this could be attributed to thermal distortion of its structure.
According to Sharon et al. (1998), the changes in pH affect the protein structure and the enzyme activity. The maximum activity of lipase from raphia mesocarp was observed to be at pH 7 which dropped onwards, this is similar to results of lipase from germinating Jatropha curcas L. seed whose pH optima was near neutrality (Abigor et al., 2003), Pentaclethra lipase which was also near neutrality (Enujiugha et al., 2004) and partial purified cocoa lipase which had an optimum pH of 7.0 (Ratna, 2008).
Metal ions have been reported to stimulate lipase-catalyzed hydrolysis of oil by removing the fatty acids from the oil-water interface and allowing lipase to act freely on oil molecules (Ohnishi et al., 1994). From this study of the effects of metallic salts on lipase activity from raphia mesocarp, all the metal salts used inhibited the lipase activity at differing levels of inhibition. The highest level of inhibition was observed with Zn2+ while the least inhibition was observed with Mn2+. Other salts used in this experiment inhibited the activity of the lipase enzyme. Monnet et al. (2012) reported that calcium ion was found to enhance lipase activity, while Mn2+, Na+, K+, Fe3+, Fe2+ and Zn2+ strongly inhibited the same. Sharon et al. (1998) reported a lipase of P. aeruginosa KKA-5 that retained its activity in presence of Ca2+ and Mg2+ but was slightly inhibited by Mn2+, Cd2+, and Cu2+. Rahman et al. (2005) stated that metal ions will bind to the enzyme and change the enzyme’s conformation to achieve better stability and hence greater activity. EDTA was also observed to inhibit the activity of the lipase and this is in agreement with the report of Monnet et al. (2012) that lipase from dormant ripe and unripe Terminalia catappaLinn were strongly inhibited by EDTA, thus suggesting that the lipases are metalloenzymes. According to Enujiugha et al.(2004), the inhibition could be attributed to its chelating process of the system and thereby disrupting the formation of the enzyme substrate complex. This invariably affects the formation of the end product (Enujiugha et al., 2004).
The result of the lipolysis activity shows that the enzyme hydrolyses coconut oil at a higher rate than other oils used. Palm oil and palm kernel oil were hydrolysed at comparable rates; also olive oil and raphia oil were hydrolysed at comparable rates. The result shows that the lipase hydrolyses medium chain triacylglycerol at a faster rate than long chain triacylglycerol. This is in contrast to the findings of Abigor et al. (2003), which reported that lipase from germinating J. curcas L. seeds hydrolyzed long-chain TAG than medium chain triacylglycerol.
In conclusion, apart from the fact that this work established the presence of lipase from Raphia mesocarp, it also revealed that this enzyme from these sources had some physico-chemical properties that are similar to what have been obtained from other plant and microbial sources. Some of these properties could make the enzyme to find extensive applications not only in chemical, pharmaceutical, food, or leather industries but also in biodegradation of some materials.
Authors declare no conflict of interest.
REFERENCES
|
Abigor DR, Opute FI, Opoku AR, Osagie AU (1985). Partial purification and some properties of the lipase present in oil palm (Elaeis gunenis Jacq.) mesocarp. J. Sci. Food Agric. 36:599-605.
Crossref
|
|
|
|
Abigor RD (2002). Production of biodiesel fuel from vegetable oils using lipases from Pseudomonas cepacia and Jatropha curcas L. Ph.D. Thesis, University of Benin, Benin city, Nigeria. p.234.
|
|
|
|
|
Abigor RD, Uadia, PO, Foglia, TA, Haas, MJ, Scott K and Savary, BJ (2003). Partial purification and properties of lipase from germinating seeds of Jatropha cucas. J. Am. Oil Chem. Soc. 79(11):1123-1128
Crossref
|
|
|
|
|
Boland W, Frobl C, Lorenz M (1991). Estrolytic and lipolytic enzymes in organic synthesis. Synthesis 12:1049-1072.
Crossref
|
|
|
|
|
Doonan S (1996). Protein purification protocols. Method Mol. Biol. 59:135-140.
Crossref
|
|
|
|
|
Enujiugha VN, Thani FA, Sanni JM and Abigor RD (2004). Lipase activity in dormant seeds of African oil bean (Pentaclethra macrophylla Benth). Food Chem. 88(3):405-408.
Crossref
|
|
|
|
|
Forgia TA, Conkerton EJ, Philip E (1995). Regio selective analysis of triacylglycerols by lipase enzymes with attractive applications for stepwise hydrolysis of triolein using Candida rugosa lipase in biphasic oil-water system. Angew. Chem. Int. Ed. 37:1608-1633.
|
|
|
|
|
Freire GDM, Castilho FL (2008). Lipases em Biocatalise in: Bon et al.(org). Enyimas em biotechnologia: Producao Applicacao e Mercado, Rio d Janeiro, Interciencia. 1:369-385.
|
|
|
|
|
Gandhi NN (1997). Applications of lipase. J. Am Oil Chem. Soc. 74:621-634.
Crossref
|
|
|
|
|
Guit RP, Kloosterman M, Menderma GW, Mayer M, Meijer EM (1991). Lipase kinetics: Hydrolysis of triacetin by lipase from Candida cylindracea in a hollow-fiber membrane reactor. Biotechnol. Bioeng. 38:727-732
Crossref
|
|
|
|
|
Henderson J, Osborne DJ (1991). Lipase activity in ripening and mature fruit of the oil palm. Stability in vivo and in vitro. Phytochemistry 30:1073-1078.
Crossref
|
|
|
|
|
Hermansyah H, Kubo M, Shibasaki-Kitakawa N, Yonemoto T (2006). Mathematical model Humicola lanuginosa lipase on its thermal stability. J. Biochem. Biophys. Acta 1547:329-338.
|
|
|
|
|
Ibrahim CO (1996). Lipolytic enzyme technology for the palm oil industry proceedings of 1996 PORIM International Oil Congress-Chemistry, Technology, Soap and Detergent Industry Conference, 23-28 Sept., 1996 at hotel Isana, Kuala Lumpur, Malaysia. pp.133-149.
|
|
|
|
|
Iftikhar T, Niaz M, ZiaMA, Haq IU (2010c). Production of extracellular lipases by Rhizopus oligosporus in a stirred fermentor. Braz. J. Microbiol. 41(4):1124-1132.
Crossref
|
|
|
|
|
Khan MY, Dahot MV and Noomrio M H (1991). Investigation of lipase activity from Cajanus cajan L seed pack. J. Sci. Ind. Res. 34:384-386.
|
|
|
|
|
Laemmli UK (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685.
Crossref
|
|
|
|
|
Lineweaver H, Burk D (1934). "The determination of enzyme dissociation constants". J. Am. Chem. Soc. 56(3):658-666.
Crossref
|
|
|
|
|
Lotrakul P, Dharmsthiti S (1997). Lipase production by Aeromonassobria LP004 in a medium containing whey and soybean meal. World. J. Mic. Biotechnol.13:163-166.
Crossref
|
|
|
|
|
Lotschert W, Beese G (1983). Collins Guide to Tropical Plants. London: Collins. ISBN 978-0-00-219112-8. 83-1496.
|
|
|
|
|
Lowry O, Rosebrough NJ, Farr AL (1951). Folin- phenol reagent. J. Biol. Chem. 193:265-275
PMid:14907713
|
|
|
|
|
Monnet TY, Pamphile KBK, Soro RY, Due EA, KouameLP (2012). Biochemical characterization of lipase activities in dormant seeds of T. catappa. J. Appl. Biosci. 24:337-3382.
|
|
|
|
|
Ohnishi K, Yoshida Y, Toita J and Sekiguchi J (1994). Purification and characterization of a novel lipolytic enzyme from Aspergillus oryzae. J. Ferm. Bioeng. 78(6):413-419.
Crossref
|
|
|
|
|
Pancreac'h G and Baratti JC (1996). Hydrolysis of p-nitrophenolpamitate in n-heptane by Pseudomonas cepacia lipase: a simple test for the determination of lipase activity in organic media. Enzyme Microb. Technol. 18:412-422.
|
|
|
|
|
Rahman R, Baharum SN, Basri M, Salleh AB(2005). High-yield purification of an organic solvent-tolerant lipase from Pseudomonas sp. strain S5. Anal. Biochem. 341(2):267-274.
Crossref
|
|
|
|
|
Ratna AS (2008). Partial characterization of llipase from cocoa beans (Theobroma cacao. L.) of clone PBC 159. Ind. J. Chem. 8(3):448-453.
|
|