Full Length Research Paper
ABSTRACT
In this study, 19 cheese samples from six common Egyptian cheese types were evaluated for microbiological safety and bacterial diversity using 16S rRNA gene amplicon sequencing analysis. Results showed that the total viable count exceeded 5.2 log CFU g-1 in 13 cheese samples. Further, the count of Enterobacteriaceae varied in the range 2.0 to 6.7 log CFU g-1 in 17 samples. PCR-based detection of pathogenic strains confirmed the absence of Listeria spp. and Salmonella spp. Furthermore, 16S rRNA gene amplicon sequencing analysis revealed the richness and diversity of the bacterial communities inhabiting the Egyptian cheese samples. Besides unfavorable bacteria, such as Enterobacteriaceae, 10 genera were present in 74% of the examined samples, predominantly including lactic acid bacteria and salt-tolerant bacteria. These insightful findings provide a deeper understanding of the typical bacterial profiles of different Egyptian cheese types and can be applied to standardize cheese production and improve cheese quality and safety.
Key words: Egyptian cheese, microbiota, food safety, fermentation product, Amplicon sequencing.
INTRODUCTION
Based on cheese processing-related archeological evidence, it is believed that Egypt, as the heart of the Middle East, was the origin and first provider of cheese. This has not only been illustrated by old Egyptian drawings (El-Gendy, 1983), but has also been proven by a recent discovery of the oldest cheese material (Greco et al., 2018). In addition to its historical significance, Egyptian cheese is also of great importance from economic, social, nutritional, and food safety perspectives. Presently, Egypt is ranked as one of the top two cheese-producing and consuming countries in Africa and the Middle East. Further, in Egypt, cheese, owing to its pleasant taste as well as its high protein, fat, and mineral contents, is a main part of meals, including lunch in schools (PM Food & Dairy Consulting, 2016).
Traditional cheeses, including soft and hard types, are primarily obtained from cow and buffalo milk. Specifically, hard-type Romy cheese, also known as Ras cheese, is produced in the shape of wheels following processing steps similar to those employed in the production of Gouda cheese, except for a few differences, especially in the subsequent dry salting step (for 4 to 5 weeks) following cheese brining, which gives Ras cheese its distinct sharp and salty flavor (Ismail et al., 2014). Further, soft traditional Egyptian cheeses include Karish, Mish, Domiati, Tallaga and Istanbuli cheeses. Specifically, Karish cheese is a fresh, healthy, and defatted cheese that is processed, usually using traditional methods, without any ripening step. Furthermore, it is used for the production of a yellowish-brown fermented cheese known as Mish cheese via pickling in microaerophilic conditions, with the addition of salt, milk, nutritive substances, oriental spices, and some plants, using old Mish cheese as the starter. This is usually followed by storage in warm conditions for a period of approximately one year to ensure ripening. Thus, Mish cheese with a sharp, salty, and pungent taste is obtained (Ismail et al., 2014).
Domiati cheese, the most popular white brined cheese in Egypt and other Arab countries that shows close resemblance with Feta cheese in Greece, is known for its unique processing technique that involves “milk salting,” during which 5 to 14% salt is directly added to the milk at the initial production step. The cheese obtained may be eaten fresh or as is often the case, the salted whey is removed, and the cheese is further stored for 4 to 8 months for ripening. Additionally, Tallaga Cheese with a creamy, spreadable soft texture and a minimal salty taste is obtained by the same method if low salt is added. Finally, to obtain Istanbuli cheese, which is characterized by a crumbly texture and Jalapeno chili studding that gives it a remarkable spicy flavor, the salt starter is added to pasteurized milk before culturing (Ismail et al., 2014).
Generally, the safety of cheese, which was previously considered as a potentially hazardous food (PHF) that facilitates microbial growth, especially when good hygiene and safety practices are not followed during its preparation, is considered to be time/temperature control (TCS) dependent. Outbreaks related to the consumption of dairy products were reported all over the world, so it is necessary to verify the safety of cheese in order to achieve the safer diet (Costard et al., 2017; Makino et al., 2005; Lance et al., 2005; MacDonald et al., 2005). It has been suggested that traditional Egyptian cheeses do not comply with both Egyptian and international cheese safety standards (Sayed et al., 2011). Various pathogenic bacteria, including Escherichia coli, Salmonella spp., Staphylococcus aureus, Bacillus cereus, and Listeria spp., which are possible sources of human infection and food poisoning, have been isolated from traditional Egyptian cheeses (El-Eyiby, 2017; Hassan and Goma, 2016). In particular, Salmonella spp. and L. monocytogenes are relatively resistant to osmotic pressure and acid stress, and can survive and proliferate during child storage (Melo et al., 2015; Leyer and Johnson, 1992). Cheese is a ready-to-eat food that is eaten without heating, so if these harmful microorganisms are contaminated in cheese, it may cause serious food poisoning.
Even though current industrial development standards for the commercial production of cheeses require milk pasteurization, most traditional Egyptian cheeses are made without pasteurization (Ismail et al., 2014) since it is well-known that these traditional practices contribute significantly to the original sensory characteristics of cheese, in addition to the associated health benefits (Montel et al., 2014). Cheeses made with raw milk are increasingly preferred by consumers due to their richer and more varied flavors than cheeses made with pasteurized milk (Yoon et al., 2016; Casalta et al., 2009). The development of cheese flavor is largely due to the naturally occurring microbial community, primarily in raw milk (Montel et al., 2014). However, such practices may be a threat to public health. It has also been reported that traditional cheeses are characterized by microbial community richness and diversity. Microorganisms such as lactic acid bacteria involved in cheese fermentation also contribute to suppressing the growth of pathogenic bacteria by producing bacteriocins and organic acids during cheese production (Montel et al., 2014; Yoon et al., 2016). Therefore, understanding the microbial flora in cheese is essential for producing safer cheese. However, analyzing these bacterial communities using culture- dependent methods is still challenging.
In this regard, the importance of using culture- independent methods, particularly 16S rRNA gene amplicon sequencing, which is based on the use of next-generation sequencing (NGS) technology, has been highlighted in several food science-related studies. Specifically, 16S rRNA gene amplicon sequencing analysis has been used for the characterization of the microbial communities in Danish raw milk cheeses (Masoud et al., 2011), the Polish cheese, Oscypek (Alegría et al., 2012), water buffalo mozzarella (Ercolini et al., 2012), artisanal Irish cheeses (Quigley et al., 2012), Latin-style cheeses (Lusk et al., 2012), and Iranian Liqvan cheese (Ramezani et al., 2017). However, as far as we know, this method has not been previously applied to characterize traditional Egyptian cheeses.
Therefore, the objective of this study was to assess the microbiological safety and biodiversity of the bacterial communities in six types of traditional Egyptian cheeses (Karish, Mish, Domiati, Tallaga, Istanbuli, and Romy cheeses) using both culture-dependent and culture- independent methods, particularly, NGS technology.
MATERIALS AND METHODS
Cheese samples
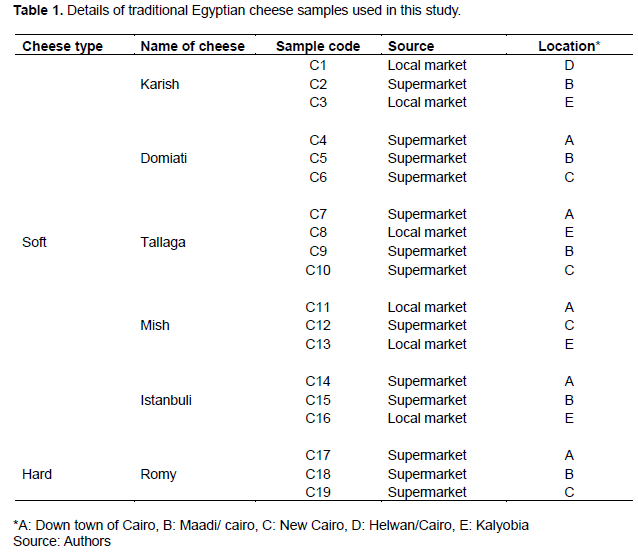
A total of 19 different commercial cheese samples corresponding to six traditional Egyptian cheese types were obtained from the Cairo and Kalyobia governorates in Egypt. Thereafter, the samples were categorized under two major groups: Soft cheese samples (Karish, Domiati, Tallaga, Mish, and Istanbuli cheeses, samples C1-C3, C4-C6, C7-C10, C11-C13 and C14-C16, respectively) and hard cheese samples (Romy cheese, samples C17-C19). After transportation to the laboratory, the cheese samples were stored at 4°C until analysis. Detailed information regarding the cheese types as well as the markets from which they were obtained are presented in Table 1.
Measurement of cheese salt concentration
Salt concentration was measured using a digital salt meter (LAQUA Twin Salt-22, Horiba, Kyoto, Japan). Specifically, 10 g of each cheese sample was homogenized with 10 mL of distilled water in a stomacher bag. Thereafter, 1 mL of each homogenized cheese sample was used for the salt concentration measurement.
Enumeration of viable bacterial counts
In a stomacher bag, 10 g of each cheese sample was homogenized with 90 mL of 0.85% saline solution for 30 s and then serially diluted. Thereafter, 1 mL of each dilution series was inoculated onto appropriate Petrifilm count plates (3 M, St. Paul, MN, USA), and to obtain the total viable cell (TVC) and Enterobacteriaceae (EB) counts, 3M aerobic count (AC) plates and Enterobacteriaceae count plates (EB/VRBG) were used, respectively. Specifically, the plates were incubated at 37°C and viable bacteria were counted after 24 and 48 h for EB/VRBG and AC, respectively. Further, De Man Rogosa Sharp Media (MRS) agar plates (MRS, Oxoid, Hampshire, UK) were used to count lactic acid bacteria (LAB) using the spread-plate method after incubation for 72 h at 30°C.
Detection of pathogenic bacteria (Listeria spp. and Salmonella spp.)
Listeria spp. were isolated on PALCAM agar media (Merck KGaA, Darmstadt, Germany) in accordance with the ISO 11290 method with modifications. Briefly, 10 g of each cheese sample was primarily enriched in 90 mL of half-Fraser broth (Oxoid) and incubated at 30°C for 24 h. This was followed by the transfer of 1 mL of this enriched solution into a 9-mL Fraser broth (Oxoid) and incubation at 30°C for another 24 h. The cultures were then plated on PALCAM agar solid media using spread-plate and streaking methods. Further, the plates were incubated at 30°C and examined after 48 h. For the detection of Salmonella spp., 10 g of each cheese sample was homogenized in 90 mL of Trypticase Soy Broth “TSB” (BD, Becton Dickinson, NJ, USA) as a pre-enrichment media and incubated overnight at 37°C. Thereafter, 1 mL of the enriched broth was transferred into a 9-mL Hajna Tetrathionate Broth (EIKEN, Tokyo, Japan) and incubated overnight at 37°C. Finally, the samples were streaked on deoxycholate hydrogen sulfide lactose agar (DHL) media (EIKEN) and incubated again at 37°C for 24 h.
Suspected colonies were then sub-cultured on the specified solid media for each strain under the same incubation conditions. Thereafter, single colonies were picked, purified, pelleted, and used for DNA extraction, which was performed using a Nucleo Spin DNA extraction kit (Macherey-Nagel, Düren, Germany) according to the manufacturer’s instructions. Additionally, for the specific detection of Listeria spp. and Salmonella spp., a designed pair of forward and reverse primers, as described by Phraephaisarn et al. (2017) and a set of primers described by Rahn et al. (1992) were used, respectively. The PCR reaction was then performed in a GeneAmp thermal cycler (PCR System 9700, Applied Biosystems, Foster City, CA, USA). For Listeria spp., amplification was conducted for 40 cycles under the following conditions: 95°C for 5 min, 95°C for 10 s, 60°C for 30 s, 72°C for 30 s, and a final extension step at 72°C for 3 min. For Salmonella spp., 30 amplification cycles were run under the following cycling conditions: 95°C for 5 min, 95°C for 15 s, 65°C for 45 s, and 72°C for 20 s using a GeneAmp thermal cycler.
Additionally, to validate the results corresponding to the examined strains, 5-µL aliquots of the PCR products were further analyzed via agarose gel electrophoresis.
16S rRNA gene amplicon sequencing analysis
To perform amplicon sequencing, 10 g of each cheese sample was added to 90 mL of 0.85% saline solution and homogenized for 30 s. Thereafter, 1-mL homogenized cheese samples were transferred into sterile Eppendorf tubes and centrifugation was performed at 15,000 ×g for 3 min. After this step, the supernatant was discarded, and the pellets obtained were stored at -20°C for microbiota analysis. For efficient bacterial DNA extraction, the obtained pellets were first treated with 100 µL of Achromopeptidase enzyme (Wako Pure Chemical Industries, Osaka, Japan), and the mixtures obtained were incubated at 55°C for 15 min. Finally, DNA extraction was performed using the NucleoSpin Tissue kit (Macherey-Nagel) in accordance with the manufacturer’s protocol. Thereafter, to obtain 2×300 bp paired-end sequences, amplicon sequencing analysis using the 16S rRNA V3/V4 region was performed using the Illumina MiSeq platform (Illumina, San Diego, CA, USA). All the analyses were outsourced to Bioengineering Lab. Co. Ltd. (Sagamihara City, Japan).
Data analysis
After amplicon sequencing, chimeric and noise sequences were removed, and representative sequences and operational taxonomic units (OTUs) were obtained. Further, to realize phylogenetic estimation, the feature-classifier plug-in (Greengene software version 13_8), which offers the possibility to compare representative sequences, was used for OTU identification at 97% similarity. Alpha diversities of the bacterial microflora in each sample were expressed using Shannon diversity index. PCA was conducted on OTUs and the calculations for PCA were done using the R “prcomp” function and plotted using the “plot” function.
RESULTS AND DISCUSSION
Physicochemical and microbiological analysis of cheese samples
In this study, the microbiological properties and salt concentrations corresponding to the 19 examined cheese samples, representing six different traditional Egyptian cheese types, including soft and hard cheeses, collected from different markets, were initially determined using conventional methods (culture-dependent methods).
As shown in Table 2, the different cheese types showed different salt concentrations, which varied from 0.5% (Karish cheese C2) to 12% (Domiati cheese C4). According to a previous study, salt concentration is a common parameter used to distinguish different cheese types (Hammam et al. 2020).
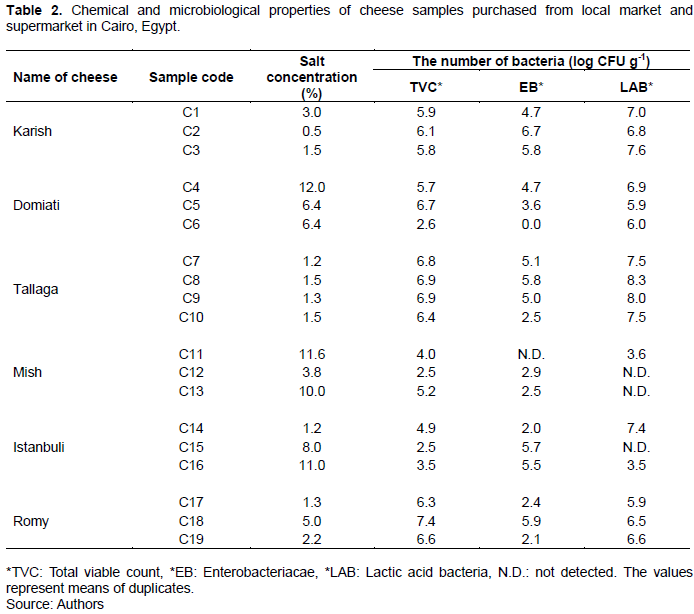
Further, the EB counts showed high variability, ranging from 2.0 (sample C14) to 6.7 (sample C2) log CFU g-1 in 17 samples, while two samples (samples C6 and C11) were completely EB free. Furthermore, the EB count observed in this study was approximately two-fold greater than that reported for Karish cheese in a previous study, which showed a maximum EB count of 3.5 log CFU g-1 (Awad, 2016). However, consistent with the results, in other studies involving Egyptian cheese samples, members of the EB group showed high counts (El-Etriby, 2017; Hassan and Gomaa, 2016; Heikal and Al-wakeel, 2014). Overall, this high range of EB count reflects a poor good manufacturing practice (GMP) from farm to fork, particularly, poor personal hygiene with possible fecal contamination. Further, this could also be attributed to the use of unpasteurized milk, which contains bacteria that may cause gastrointestinal illness as well as cheese spoilage (Baylis et al., 2011). Moreover, this high bacterial count possibly contributes to cheese defects, such as color change, gas production, and hole formation, which should not be ignored (Roberts et al., 1998).
In this study, 16 out of the 19 cheese samples showed high lactic acid bacteria (LAB) counts, in the range 3.5 to 8.3 log CFU g-1, whereas the three remaining samples were LAB free. In a previous study, an average LAB count of 7.8 log CFU g-1 was reported for Egyptian cheese (Awad, 2016). In another study, different cheese types showed different LAB counts, with the maximum value (7.4 log CFU g-1) corresponding to Domiati cheese (El-Baradei et al., 2007). Reportedly, LAB, which are responsible for the organoleptic characteristics of cheeses, show health benefits via various mechanisms owing to their probiotic nature. The high LAB count in Egyptian cheeses, which is also frequently observed in fermented products, possibly plays an important role in the control of pathogenic bacteria, such as Listeria spp. and Salmonella spp. (Losito et al., 2014). In this study, based on culturing on their specific media, eight and four typical of Listeria spp. and Salmonella spp. colonies were observed, respectively; however, PCR-based analyses confirmed their absence in all the examined samples (data not shown). The negative PCR-based results for these two species contradict the results of previous studies, which reported the presence of pathogenic bacteria, including Listeria spp. (Ismail et al., 2014) and Salmonella spp. in Egyptian cheese (El-Baz et al., 2017). This implies that, in some cases, the use of traditional culture-dependent methods may generate misleading results given the possibility of the co-existence of microorganisms with similar growth conditions. Thus, to obtain accurate results, culture-independent PCR analysis is required.
Bacterial diversity of cheeses
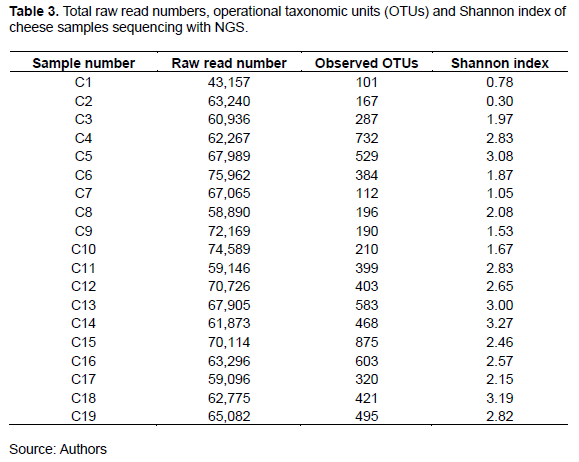
To determine the bacterial communities, cheese samples were analyzed via 16S rRNA gene amplicon sequencing. Thus, large numbers of sequence reads with similarity were clustered into OTUs. As shown in Table 3, the observed OTUs, read numbers, and Shannon diversity indices varied among the cheese types. Specifically, the lowest OTU number (101) corresponded to Karish cheese sample C1, while the highest recorded value (875) corresponded to Istanbuli cheese sample C15. Further, the Shannon diversity indices (mean = 2.3), which ranged from 0.30 (sample C2) to 3.27 (sample C14), indicated the presence of a diverse microbiota in the Egyptian traditional cheese samples. These findings contradict those of some previous studies, which showed only moderate variation between the diversity indices corresponding to different cheese types (Quigley et al., 2012; Riquelme et al., 2015).
Overall comparison of each cheese types
In this study, 25 out of the 70 detected microbiota families had relative abundances above 1% in at least one of the cheese samples. The most abundant families, which showed varied relative abundances in the examined cheese samples, included Streptococcaceae, Lactobacillaceae, Leuconostocaceae, Enterococcoceae, Staphylococcaceae, Xanthomonodaceae, Pseudomonadaceae, Oceanospirrillaceae, Vibrionaceae and Enterobacteriaceae (data not shown). Minor families, such as Bifidobacteriaceae were also frequently observed. Thus, the Egyptian cheese samples seemingly contain a higher abundance of bacterial families than Italian (Marino et al., 2017) and Pico Portuguese (Riquelme et al., 2015) cheese samples. Moreover, Bifidobacteriaceae, which reportedly, cannot survive at salt concentrations ≥5% (De Castro-Cislaghi et al., 2012), was observed in four of the Egyptian cheese samples with high salt concentrations (6.4 to 12%), suggesting that some of the strains had developed high-salinity tolerance. This also suggests the possibility for their application as probiotic candidates for foods with high salt contents, such as cheese (Alegría et al., 2012; Marino et al., 2017).
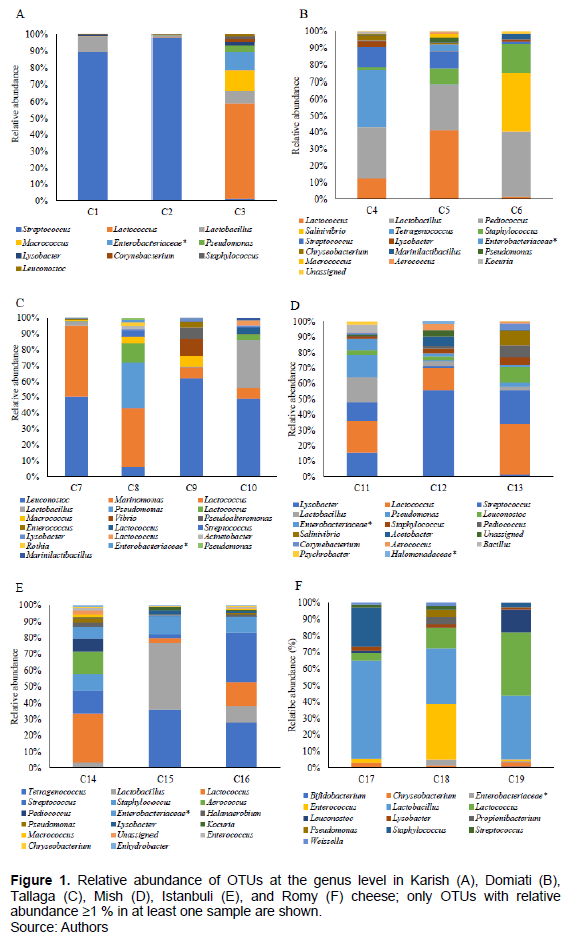
Considering a lower taxonomic level, a total of 101 different genera were identified, 39 of which were present at 1% relative abundance in at least one cheese sample (Figure 1A to E). Further, the results indicated that 10 genera (Streptococcus, Lactococcus, Lactobacillus, Leuconostoc, Enterococcus, Staphylococcus, Lysobacter, Enterobacteriaceae, Pseudomonas, and Acinetobacter) constituted the core microbiome of the cheese samples. They were observed in at least 14 of the 19 examined cheese samples even though their proportions varied between samples (the Karish cheese samples did not show the presence of Enterococcus). Furthermore, the first five genera of LAB constituted the majority of the core microbiota in the cheese samples, indicating that they are responsible for the distinctive taste of the cheese samples, despite their varying relative abundances among the different cheese types. Unfortunately, the other five observed LAB are considered to be problematic in cheese environments. For example, consistent with our microbiological count results, the presence of Enterobacteriaceae is indicative of poor hygienic conditions (Baylis et al., 2011). Additionally, Pseudomonas, which usually results from the use of raw milk for cheese production, is responsible for various kinds of cheese defects and spoilage, as well as biofilm formation (Johnson and Sommer, 2020). In this study, Macrococcus and Chryseobacterium were also frequently detected; their presence was confirmed in 12 out of 19 examined cheese samples. Minor genera, including Tetragenococcus, were also observed in three of the cheese samples examined in this study (relative abundances in the range 28 to 29.6%). Further, given that this study is the first, to the best of the researchers’ knowledge, in which Egyptian cheese samples are analyzed using a robust technique; the presence of several genera, such as Lysobacter, Chryseobacterium, Marinilactibacillus, Weissella, Enhydrobacter, Planococcaceae, Tetragenococcus, and Halanaerobium was reported, in Egyptian cheese samples for the first time. Interestingly, four genera that have not been previously associated with cheese were observed in this study. These included Lysobacter, which constituted part of the core microbial composition of all the 19 examined samples, as well as Salinivibrio, Alteromondales and Candidatus.
The relative abundances of different bacteria genera showed obvious variations among the different cheese types, and in some cases, varied between samples of the same cheese type. Specifically, Lactobacillus spp. and Enterococcus species were more prevalent in Romy cheese samples (hard cheese); however, their relative abundances in soft cheese samples were much lower. Streptococcus showed a higher relative abundance in Karish cheese samples; however, its relative abundance in Tallaga cheese samples was lower than that observed in the other cheese samples. Additionally, Lactococcus species showed greater predominance in all the five soft cheese samples than in the hard one. Our results also indicated that Enterobacteriaceae were widespread in all the cheese types, showing a higher relative abundance in Karish cheese, which is a soft-type cheese (sample C3; 11.8%). Further, Streptococcus spp. showed a high relative abundance in Karish cheese samples C1 and C2 (88.9 and 96.2%, respectively). The result also confirmed the presence of Tetragenococcus in Domiati and Istanbuli cheese samples, while Leuconostoc and Marinomonas showed very high relative abundances in Tallaga cheese samples.
Karish cheese
Considering the microbial communities in the individual cheese samples observed in this study, it was evident that Karish cheese samples predominantly contained LAB, including Streptococcus, Lactobacillus, and Lactococcus (Figure 1A), which were also the major part of the bacterial profile obtained via the TTGE analysis of Karish cheese (El-Baradei et al., 2005). This is consistent with the findings corresponding to raw milk cheese, such as Danish cheese (Masoud et al., 2011). Further, in this study, Streptococcus showed distinctive abundance and dominance in two of the Karish cheese samples, C1 and C2 (88.9 and 96.2%, respectively), while Lactococcus showed predominance in the third Karish cheese sample (sample C3, 53.1%). Thus, Streptococcus possibly contributes more to the fermentation of Karish cheese than Lactobacillus and Lactococcus species. Interestingly, in sample C3, with a lower Streptococcus relative abundance (2.2%), a high abundance of Staphylococcaceae (13.8%), including Staphylococcus and Macrococcus, was observed, suggesting that either Streptococcus is not a good competitor with Staphylococcaceae (Janek et al., 2016), or that the three Karish samples were not produced under the same standardized conditions possibly owing to the starter cultures that were used (Hammam et al., 2020).
Domiati cheese
In this study, Domiati cheese showed a complex and unique profile that reflected extensive diversity (Figure 1B). Generally, Domiati cheese was dominated by LAB genera (that is, Streptococcus, Lactococcus, Lactobacillus, Tetragenococcus and Pediococcus) as well as non-LAB genera (Staphylococcus and Salinivibrio). However, the three Domiati cheese samples showed differences in the dominant bacteria. Specifically, in sample C4, Tetragenococcus, Lactobacillus, Streptococcus, and Lactococcus were dominant (29.4, 26.9, 11.2 and 7.9%, respectively). Further, in sample C5, Lactococcus, Lactobacillus, Streptococcus, and Staphylococcus were dominant (31.6, 19.2, 9.2 and 7.7%, respectively), and in sample C6, Pediococcus, Salinivibrio, and Staphylococcus were dominant (37, 33.3 and 16.8%, respectively). The first study of the bacterial profile of Domiati cheese via TTGE and DGGE suggested that its bacterial content can be classified under three main groups, namely, dominant, frequently encountered, and occasionally encountered bacterial species (El-Baradei et al., 2007), and in this study, all these three main groups, with different species categories was basically observed. Furthermore, via NGS, new genera in Domiati cheese that had not been previously identified were observed. Some of these newly reported genera (Lysobacter, Pseudomonas, Chryseobacterium, and Marinilactibacillus) were present in the three Domiati cheese samples; another (Tetragenococcus) was present in two of the Domiati cheese samples, while two others (Salinivibrio and Alteromondales) were present in one of the Domiati cheese samples. Most likely, Tetragenococcus spp., which showed predominance in sample C4, with the highest salt concentration (12%), considering all the six cheese types, survived owing to its halotolerant nature. Further, this bacterial genus, together with Pediococcus spp., which was also observed in Domiati cheese samples, could confer possible health benefits owing to their probiotic function (Marino et al., 2017). In accordance with a previous study (El-Baradei et al., 2007), we could conclude that the microbiome of most of the Domiati cheese samples consisted of salt-tolerant and marine bacteria, owing to their high salt contents, which probably contributed to the ripening process of the Domiati cheese samples.
Tallaga cheese
Regarding the Tallaga cheese samples, three (C7, C9, and C10) predominantly consisted of Leuconostoc spp. (50.1, 60.9 and 47.1%, respectively), followed by Marinomonas in sample C7 (43.9%), Vibrio in sample C9 (10.3%), and Lactobacillaceae in sample C10 (29.3%). Conversely, Tallaga cheese sample C8 showed a different dominance pattern with Lactococcus showing predominance (46.1%), followed by Pseudomonas (29.6%) (Figure 1C). Interestingly, considering all the cheese types examined in this study, Marinomonas and Vibrio were only observed in the Tallaga cheese samples. Specifically, Marinomonas was identified as one of the dominant bacteria in sample C7; however, its relative abundances in the other Tallaga cheese samples (samples C9 and C10) were lower (6.6%), while Vibrio represented 10.3% of the total genera in sample C9 and 0.1% in sample C10. Based on these findings regarding the bacterial ecosystem in Tallaga cheese, which we report for the first time, it could be concluded that the process of Tallaga cheese manufacturing is primarily a Leuconostoc-driven process, with contributions from other LAB. Findings also suggested that except for two genera (Marinomonas and Bacillus), the bacterial profile of Tallaga cheese samples was part of that of Domiati cheese samples; this may be attributed to the similarities between their manufacturing process (except for salinity and ripening time) (Ismail et al., 2014) Nevertheless, these two cheese types showed obvious differences with respect to the survival of some salt-tolerant bacteria, such as Tetragenococcus, Corynebacterium, Kocuria and Salinivibrio in the high-salt-containing Domiati cheese samples.
Mish cheese
Considering all the examined Egyptian cheese samples, Mish cheese appeared to have the highest bacterial diversity given that it harbored almost all the detected bacterial genera so far reported in this study (except Marinomonas and Vibrio) (Figure 1D). Specifically, the bacterial profile of the Mish cheese samples was predominated by LAB, and genera such as Lactococcus, Streptococcus, Lactobacillus, Leuconostoc, and Enterococcus, with different abundances, were detected in all the samples. Further, other genera, including LAB and non-LAB were detected in two and one samples, respectively. In particular, sample C11 was dominated by Lactococcus spp. (18.6%), Lactobacillus (14.9%), non-LAB (Lysobacter and Pseudomonas, 14.1 and 13%, respectively), and Streptococcus (11.1%). Other subdominant bacterial genera included Entero- bacteriaceae, Bacillus, Leuconostoc, Corynebacterium, Staphylococcus, and Psychrobacter (7.7, 4.6, 3.0, 2.7, 2.0 and 2.0%, respectively). Furthermore, in sample C12, Lysobacter spp. was predominant and showed its highest relative abundance (46.7%), considering all the 19 cheese samples examined in this study; the next abundant genera were Lactococcus (10.3%) and Acetobacter (5.9%). Additionally, bacterial genera, including Lactococcus, Streptococcus, Leuconostoc, and Salinivibrio showed predominance in sample C13 (27.7, 21.3, 8.7 and 8.1%, respectively), followed by Pediococcus (6.7%), Corynebacterium (5.2%), and Staphylococcus (4.9%), and given that Mish cheese is primarily made from Karish cheese, it contained all the bacterial genera observed in the Karish cheese samples. LAB was also identified as the dominant bacteria that possibly played a primary functional role in Mish cheese ripening owing to the favorable microaerophilic conditions as well as the high salt concentration, even though this was not consistent with the results obtained using the culture-dependent method, which did not show LAB growth (Table 2). LAB either entered the viable but not-culturable state (Millet and Lonvaud-Funel, 2000) or most probably were affected by acid stress, which is a self-imposed stress that leads to cell death owing to prolonged exposure to an acidic environment resulting from the accumulation of lactic acid by LAB during the over 1-year ripening of Mish cheese (Even et al., 2002).
Istanbuli cheese
Regarding Istanbuli cheese, a vast diversity of bacterial microbiota was reported for the first time; Lactococcus, Streptococcus, Lactobacillus, and Tetragenococcus showed high relative abundances among the identified LAB. Possibly they represented the major contributors to cheese flavor and the cheese ripening process; other LAB and non-LAB, with different relative abundances, were also frequently detected (Figure 1E). For individual samples, Lactococcus, Streptococcus, Aerococcus, and Staphylococcus were identified as the dominant genera in sample C14 (18.8, 14.4, 11.6 and 8.2%, respectively), followed by others; Streptococcaceae (7.5%), Pediococcus (6.9%), and Enterobacteriaceae (6.8%). Further, minor genera, with relative abundance below 5%, including, but not limited to Lactobacillus, Pseudomonas, Enterococcus, Macrococcus, Chryseobacteria, Acinetobacter, Marinilactibacillus, Halanaerobium, Enhydrobacter, and Alteromondalis, were also detected. In sample C15, Lactobacillus, Tetragenococcus, and Staphylococcus showed predominance (35.9, 29.6, and 17%, respectively), whereas sample C16 primarily consisted of Streptococcus (31.6%), Tetragenococcus (28%), Lactococcus (12.6%), and Lactobacillus (11.6%). Interestingly, Istanbuli cheese samples C15 and C16 showed the highest prevalence of Tetragenococcus (28-29.6%) and along with Domiati cheese sample C4, the highest prevalence of Kocuria, a minor genus (1.1-1.6%). In general, the genus level bacterial profile and diversity corresponding to Istanbuli and Domiati cheeses showed several similarities, possibly owing to their high salinities, which allowed the survival of salt-tolerant and marine bacteria. Reportedly, Kocuria spp., which have been isolated from milk and fermented dairy products in India, is associated with Domiati cheese (El-Baradei et al., 2007), and their psychrotropic and salt-resistant nature may be the reason for their growth in cheese (Patil, 2019).
Romy cheese
Contrary to the variation of bacterial communities among soft cheese samples of the same type, the hard cheese samples (Romy cheese samples) showed great similarity (Fig 1F). Specifically, Romy cheese samples C17, C18, and C19, were dominated by Lactobacillus spp. (54.8, 31.3, and 32.2%, respectively), followed by Staphylococcus (22.6%) in sample C17, Enterococcus (30.7%) in sample C18, and Streptococcaceae (19.4%), Lactococcus (16.3%), and Leuconostoc (13.3%) in sample C19. Further, minor genera, such as Pseudomonas, Enterobacteriaceae, Chryseobacterium, Weissella, and Propionibacterium, were also detected in all these three samples; however, with different relative abundances. Further, the latter two species showed more notable relative abundances in the Romy cheese samples than in the other cheese types. The results here reported are similar to those obtained after the first systematic profiling of Romy cheese using NGS, which identified Lactobacillus, Staphylococcus, Streptococcus, and Lactococcus as the dominant bacterial genera, and possibly, these are primarily responsible for the sensory attributes and ripening process of Romy cheese. Furthermore, these bacterial genera have also shown dominance in Gouda cheese, which is manufactured via a similar process. However, the proportion of Staphylococcus in Romy cheese (1.5-22.6%) was found to be slightly higher than that in Gouda cheese (2.0-13.4%). However, the relative abundance of Lactococcus in the Romy cheese samples examined in this study (1.3-16.3%) was lower than that previously observed for Gouda cheese (40.1-49.1%) (Salazar et al., 2018). Differences between the commonly used milk or starter cultures employed may explain this variation.
Differences in microbial community structure by PCA
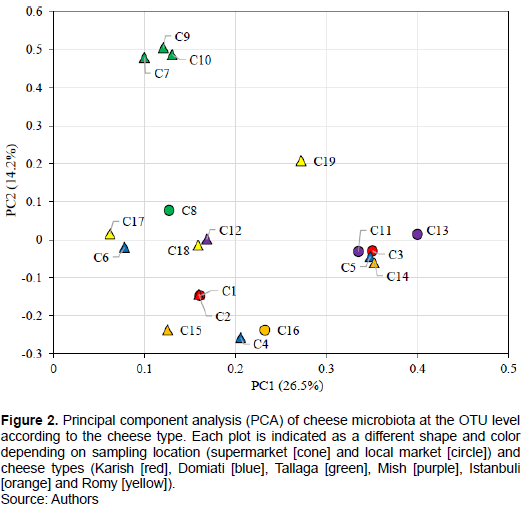
PCA was performed to investigate the possibility of clustering bacterial communities according to cheese type and salt concentration. The results thus obtained (Figure 2) indicated that most of the samples tended to show clustering according to cheese type, and the highest homology corresponded to Karish cheese samples C1 and C2, excluding sample C3. Further, Tallaga cheese samples C7, C9, and C10, excluding sample C8, all of which were obtained from local markets, showed clustering. It was observed that for Mish, Istanbuli, and Romy cheese samples, two out of three samples showed clustering with relatively lower homology compared with the Karish and Tallaga cheese samples that showed clustering. However, in the case of Domiati cheese, the three samples, C4, C5 and C6 did not show any clustering, possibly because the high salt concentration of sample C4, which was approximately double those of samples C5 and C6, might have resulted in the clustering of this sample with other high-salt-containing samples (Istanbuli cheese samples C16 and C15). Further, the three cheese samples C11, C3, and C13 which were all obtained from the local market, showed another clustering despite them originating from different cheese types. Thus, it could be suggested that three factors, cheese type, salinity, and manufacturing technique/market type, are primarily responsible for the bacterial microbiota composition of traditional Egyptian cheeses. The majority of the cheese samples, excluding Domiati cheese samples, could also be clustered based on their type. Particularly, the greatest homology was observed in low-salt-containing cheese samples (Karish and Tallaga cheese samples), while high-salt-containing Domiati cheese samples did not show any clustering based on cheese type. They rather showed clustering with other samples of different cheese types probably owing to their high salt concentration. Moreover, samples obtained from the local market showed a third clustering pattern. This finding highlights the importance of the standardization of processing techniques for the analysis of different dairy producers so that harmonized results can be obtained. In general, it is understood that no particular factor decisively determines the composition of the microflora of the complex cheese environment; a set of factors, from farm to fork, with relevance to industry have been previously discussed in this regard (Marino et al., 2017; Montel et al., 2014; Yeluri et al., 2018).
CONCLUSION
This study is the first in which the bacterial communities in traditional Egyptian cheese samples were investigated via 16S rRNA gene amplicon sequencing analysis. Based on NGS technology, various LAB known for their probiotic properties, constituted a major part of the bacterial profile of the cheese samples examined in this study. Unfortunately, the presence of pathogenic bacteria and bacteria with cheese spoilage ability was also confirmed. These results, in combination with those obtained using culture-dependent approaches, suggest a public health risk, the production of low-quality cheese with possible defects, and poor GMP. Regardless, the results of this study provide valuable information that can help cheese producers obtain specific and standardized characterization, minimize product variation, and enhanced cheese quality as well as safety.
CONFLICT OF INTERESTS
The authors have not declared any conflict of interests.
ACKNOWLEDGMENTS
The author appreciates Editage (www.editage.com) for English language editing.
REFERENCES
|
Alegría Á, Szczesny P, Mayo B, Bardowski J, Kowalczyk M (2012). Biodiversity in Oscypek, a traditional Polish Cheese, determined by culture-dependent and -independent approaches. Applied Environmental Microbiology 78(6):1890-1898. |
|
|
Awad S (2016). Microbial safety criteria and quality of traditional Egyptian Karish cheese. African Journal of Microbiological Research 10(22):804-812. |
|
|
Baylis C, Uyttendaele M, Joosten H, Davis A (2011). The Enterobacteriaceae and their significance to the food industry. In ILSI Europe Report Series pp. 1-48. |
|
|
Casalta E, Sorba JM, Aigle M, Ogier JC (2009). Diversity and dynamics of the microbial community during the manufacture of Calenzana, an artisanal Corsican cheese. International Journal of Food Microbiology 133(3):243-251. |
|
|
Costard S, Espejo L, Groenendaal H, Zagmutt FJ (2017). Outbreak-Related Disease Burden Associated with Consumption of Unpasteurized Cow's Milk and Cheese, United States, 2009-2014. Emerging Infectious Diseases 23(6):957-964. |
|
|
De Castro-Cislaghi FP, Silva CDRE, Fritzen-Freire CB, Lorenz JG, S.Sant'Annab E (2012). Bifidobacterium Bb-12 microencapsulated by spray drying with whey: Survival under simulated gastrointestinal conditions, tolerance to NaCl, and viability during storage. Journal of Food Engineering 113(2):186-193. |
|
|
El-Baradei G, Delacroix-Buchet A, Ogier JC (2007). Biodiversity of bacterial ecosystems in traditional Egyptian Domiati cheese. Applied Environmental Microbiology 73(4):1248-1255. |
|
|
El-Baradei G, Delacroix-Buchet AA, PERY P Ogier JC (2005). Identification of bacterial communities of Egyptian Karish cheese using molecular footprinting tools. Egyptian Journal of Dairy Science 33:25-34. |
|
|
El-Baz AH, El-Sherbini M, Abdelkhalek A, Al-Ashmawy MA (2017). Prevalence and molecular characterization of Salmonella serovars in milk and cheese in Mansoura city, Egypt. Journal of Advanced Veterinary and Animal Research 4(1):45-51. |
|
|
El-Etriby DE (2017). Microbiological study of some cheese and milk powder samples. Egyptian Journal of Medical Microbiology 26(1):25-32. |
|
|
El-Gendy SM (1983). Fermented foods of Egypt and the Middle East. Journal of Food Protection 46(4):358-367. |
|
|
Ercolini D, De Filippis F, La Storia A, Lacono M (2012). "Remake" by high-throughput sequencing of the microbiota involved in the production of water Buffalo mozzarella cheese. Applied Environmental Microbiology 78(22):8142-8145. |
|
|
Even S, Lindley N, Loubière P, Cocaign-Bousquet M (2002). Dynamic response of catabolic pathways to autoacidification in Lactococcus lactis: transcript profiling and stability in relation to metabolic and energetic constraints. Molecular Microbiology 45(4):1143-1152. |
|
|
Greco E, El-aguizy O, Ali M, Foti S, Cunsolo V, Saletti R, Ciliberto E (2018). Proteomic analyses on an ancient Egyptian cheese and biomolecular evidence of brucellosis. Analytical Chemistry 90(16):9673-9676. |
|
|
Hammam ARA, Elfaruk MS, Ahmed ME, Sunkesula V (2020). Characteristics and technological aspects of the Egyptian cheeses. International Journal of Current Microbiology and Applied Science 9(6):3338-3354. |
|
|
Hassan GM, Gomaa SM (2016). Microbiological quality of soft cheese marketed in Cairo and Giza governorates. Alexandria Journal of Veterinary Sciences 50(1):18-23. |
|
|
Heikal GI, Al-wakeel SA (2014). Bacteriological hazard of white cheese processed in some small rimitive lants (dairy shops) in Tanta city. Benha Veterinary Medical Journal 26(1):185-194. |
|
|
Ismail D, Rabie M, Naby RA, Adel Y, Elmiligy B, Haddad S, Bencharif A, Tozanli S, Lapujade J (2014). Developing the typical dairy products of Alexandria and Beheira diagnosis and local strategy. HAL open science. |
|
|
Janek D, Zipperer A, Kulik A, Krismer B, Peschel A (2016). High frequency and diversity of antimicrobial activities produced by nasal staphylococcus strains against bacterial competitors. PLoS Pathogen 12(8):e1005812. |
|
|
Johnson M, Sommer DC (2020). Dairy Pipeline. Wisconsin Center for Dairy Research 25:1-12. |
|
|
Lance H, Predy G, Hislop N, Chui L, Kowalewska-Grochowska K, Trottier L, Kreplin C, Zazulak I (2005). An Outbreak of E. coli O157:H7 Hemorrhagic Colitis Associated with Unpasteurized Gouda Cheese. Canadian Journal of Public Health 96(3):182-184. |
|
|
Leyer GJ, Johnson EA (1992). Acid adaptation promotes survival of Salmonella spp. in cheese. Applied and Environmental Microbiology 58(6):2075-2080. |
|
|
Losito F, Arienzo A, Bottini G, Priolisi FR, Mari A, Antonini G (2014). Microbiological safety and quality of Mozzarella cheese assessed by the microbiological survey method. Journal of Dairy Science 97(1):46-55. |
|
|
Lusk TS, Ottesen AR, White JR, Allard MW, Brown EW, Kase JA (2012). Characterization of microflora in Latin-style cheeses by next-generation sequencing technology. BMC Microbiology 12(1):1-10. |
|
|
MacDonald PDM, Whitwam RE, Boggs JD, MacCormack JN, Anderson KL, Reardon JW, Saah JR, Graves LM, Hunter SB, Sobel J (2005). Outbreak of listeriosis among Mexican immigrants as a result of consumption of illicitly produced Mexican-style cheese. Clinical Infectious Diseases 40(5):677-682. |
|
|
Makino S, Kawamoto K, Takeshi K, Okada Y, Yamasaki M, Yamamoto S, Igimi S (2005). An outbreak of food-borne listeriosis due to cheese in Japan, during 2001. International Journal of Food Microbiology 104(2):189-196. |
|
|
Marino M, Innocente N, Maifreni M, Mounier J, Cobo-Díaz J, Coton E, Carraro L, Cardazzo B (2017). Diversity within Italian cheesemaking brine-associated bacterial communities evidenced by massive parallel 16S rRNA gene tag sequencing. Frontiers in Microbiology 8:2119. |
|
|
Masoud W, Takamiya M, Vogensen FK, Lillevang S, Al-Soud WA, Sørensen SJ, Jakobsen M (2011). Characterization of bacterial populations in Danish raw milk cheeses made with different starter cultures by denaturating gradient gel electrophoresis and pyrosequencing. International Dairy Journal 21(3):142-148. |
|
|
Melo J, Andrew PW, Faleiro ML (2015). Listeria monocytogenes in cheese and the dairy environment remains a food safety challenge: The role of stress responses. Food Research International 67:75-90. |
|
|
Millet V, Lonvaud-Funel A (2000). The viable but non-culturable state of wine micro-organisms during storage. Letters in Applied Microbiology 30(2):136-141. |
|
|
Montel M, Buchin S, Mallet A, Delbes-paus C, Vuitton DA, Desmasures N, Berthier F (2014) Traditional cheeses: Rich and diverse microbiota with associated benefits. International Journal of Food Microbiology 177:136-154. |
|
|
Patil SH (2019). Psychrotrophic microbiota in milk and fermented milk products. Journal of Pure Applied Microbiology 113(2):1257-1266. |
|
|
Phraephaisarn C, Khumthong R, Takahashi H, Oshima C, Kodama K, Techaruvichit P, Vesaratchavest M, Taharnklaew R, Keeratipibul S (2017). A novel biomarker for detection of Listeria species in food processing factory. Food Control 73:1032-1038. |
|
|
Quigley L, O'Sullivan O, Beresford T, Paul Ross R, Fitzgerald GF, Cotter PD (2012). High-throughput sequencing for detection of subpopulations of bacteria not previously associated with artisanal cheeses. Applied Environmental Microbiology 78:5717-5723. |
|
|
Rahn K, Grandis SD, Clarke RC, McEwen SA, Galán JE, Ginocchio C, Curtiss 3rd R, Gyles CL (1992). Amplification of an invA gene sequence of Salmonella typhimurium by polymerase chain reaction as a specific method of detection of Salmonella. Molecular and Cellular Probes 6(4):271-279. |
|
|
Ramezani M, Hosseini SM, Ferrocino I, Amoozegar MA, Cocolin L (2017). Molecular investigation of bacterial communities during the manufacturing and ripening of semi-hard Iranian Liqvan cheese. Food Microbiology 66:64-71. |
|
|
Riquelme C, Câmara S, Enes Dapkevicius M, Vinuesa P, Costa C, da Silva G, Malcata FX, Rego AO (2015). Characterization of the bacterial biodiversity in Pico cheese (an artisanal Azorean food). International Journal of Food Microbiology 192:86-94. |
|
|
Roberts TA, Cordier JL, Gram L, Tompkin RB, Pitt JI, Gorris LGM, Swanson KMJ (2005a). Chapter 5: Vegetables and vegetable products. In: T. A. Roberts, J.-L. Cordier, L. Gram, R. B. Tompkin, J. I. Pitt, L. G. M. Gorris, & K. M. J. Swanson (Eds.), Micro-Organisms in Foods 6: Microbial Ecology of Food Commodities (2nd ed., pp. 277-325). New York: Kluwer Academic and Plenum Publishers. |
|
|
Salazar JK, Carstens CK, Ramachandran P, Shazer AG, Narula SS, Reed E, Ottesen A, Schill KM (2018). Metagenomics of pasteurized and unpasteurized gouda cheese using targeted 16S rDNA sequencing. BMC Microbiology 18:1-13. |
|
|
Sayed M, Abdel-Hameid A and Shaban W (2011). Microbiological evaluation of some Egyptian white soft cheese. Benha Veterinary Medical Journal 1:1-6. |
|
|
PM Food & Dairy Consulting (2016). World Cheese Market 2000-2023. |
|
|
Yeluri Jonnala BR, McSweeney PLH, Sheehan JJ, Cotter PD (2018). Sequencing of the Cheese microbiome and its relevance to industry. Front Microbiology 9:1020. |
|
|
Yoon Y, Lee A, Choi K (2016). Microbial benefits and risks of raw milk cheese. Food Control 63: 201-215. |
|
Copyright © 2024 Author(s) retain the copyright of this article.
This article is published under the terms of the Creative Commons Attribution License 4.0