ABSTRACT
Seven hundred and eight (708) samples collected from the goats of Gujarat were screened for Mycoplasma agalactiae by culture and PCR using 16S rRNA based genus specific and species-specific primers. Amplification of the p30 membrane protein gene was carried out using specific primers and the resultant amplicons were subjected to restriction enzyme analysis. The isolates yielded 715 bp and 360 bp products with genus-specific and species-specific primers, respectively and were identified as M. agalactiae. Amplification of the p30 membrane protein gene yielded a 730 bp product. Restriction enzyme analysis of the 730 bp amplicon of p30 gene with RsaI and MboII yielded 2 fragments (654 bp and 74 bp) and 3 fragments (111 bp, 375 bp and 244 bp), respectively. Digestion with Sau3AI yielded two fragments (461 bp and 269 bp) while digestion with AluI resulted in 3 fragments (342 bp, 328 bp and 60 bp). The results of the present study revealed the presence of polymorphism at the respective positions of p30 membrane protein gene of M. agalactiae isolates examined by the restriction digestion. These polymorphisms can result into changes in pathogenesis and persistence inside the host and require further investigation of immunological outcome of these polymorphisms.
Key words: Mycoplasma agalactiae, polymerase chain reaction (PCR), restriction enzyme, p30.
In small ruminants, Mycoplasma agalactiae is responsible for a syndrome known as contagious agalactiae of small ruminants (CASR) (Solsona et al., 1996), which is generally characterized by mastitis, arthritis and keratoconjunctivitis. It causes reduction and suppression of milk production (De Garnica et al., 2013) and occasionally results into abortion and death (Madanat et al., 2001). For pathogenesis and clinical manifestations of mycoplasmal infections, adhesion to host cells and immune evasion are the major prerequisite for colonization. Different strategies used for the successful persistence of M. agalactiae inside host include a constantly changing surface structure and the capacity of some lipoproteins to induce the expression of up- and down modulating cytokines (Razin et al., 1998). Few constantly expressed surface proteins have also been described in M. agalactiae which includes p30, p48 and p80 as immunogens and other proteins belonging to the variable surface membrane proteins family (Vpma) (Cacciotto et al., 2010). Vpma phase variation has been reported to have importance for survival and persistent infection of M. agalactiae in host (Chopra-Dewasthaly et al., 2017). The M. agalactiae variable gene (avg) system is a cluster of four genes that encode a family of surface lipoproteins characterized by high-frequency phase and size variations (Flitman-Tene et al., 2000). Stability and variation of these membrane proteins have implications for immunogenicity and pathogenicity. Among the immunodominant membrane proteins, expression of p30 was detected consistently from several strains of M. agalactiae revealing its importance for serological analysis (Fleury et al., 2001). Membrane proteins p80 (Kashoo et al., 2011) and p40 (Fleury et al., 2002) were also reported to have serological importance in the M. agalactiae infections and their importance as candidate proteins for diagnosis. Several studies have targeted these membrane protein genes for identification of the organisms (Macun et al., 2010). Among the various molecular techniques, restriction analysis is an important technique of genetic characterization and to study the variation among the strains of Mycoplasma circulating in the field. In the present study, p30 membrane protein gene was targeted to study the polymorphism among the isolates obtained from the samples collected from goats of Gujarat, which has significant number of goat and sheep population including several well-known milch and dual purpose breeds of India.
Collection and processing of samples for cultural isolation
Ear (169), nasal (158) and ocular (28) swabs as well as lung tissues (94) and milk (259) samples were collected aseptically from healthy as well as sick animals and processed for isolation of Mycoplasma. After collection, each sample was placed directly in 2 ml of MBHS-L broth (Modified Balanced Hank’s Salt Solution Liquid Media) and kept at 37°C for 1 h. After incubation for 1 h, 200 µl of each MBHS-L broth containing sample was transferred to 2 ml of fresh MBHS-L broth after filtration (with 0.45 µm filter). The fresh MBHS-L broths containing filtered inoculum were incubated at 37°C for 10-15 days. The broths were examined daily for sign of growth (floccular material) and positive cultures were further purified. The samples showing growth in broth were inoculated on solid media (Modified Balanced Hank’s Salt Solution Agar Media, MBHS-A) and incubated anaerobically for 10 days at 37ºC under humid conditions in 5-10% carbon dioxide tension to obtain optimum growth (Carmichael et al., 1972).
Examination of the isolated colonies and biochemical characterization of isolates
The suspected colonies of Mycoplasma were examined morphologically under microscope (4X) after staining them with different stains viz. Dienes’, Giemsa and Acridine orange stain. Biochemical tests as described by Erno and Stipkovits (1973) such as catabolism of glucose, hydrolysis of arginine, phosphatase activity, tetrazolium reduction, serum digestion, digitonin sensitivity test (Freundt, 1973) and film and spot formation were carried out to determine the biochemical activity of the suspected Mycoplasma isolates.
Confirmation of Mycoplasma isolates by polymerase chain reaction (PCR)
For preparation of the template DNA, 2 ml of broth culture of each isolate was centrifuged at 12000 rpm in a micro-centifuge at 4°C for 25 min. The pellets were washed in 500 ml of PBS twice; pellets were resuspended in 100 ml of nuclease-free water and boiled for 10 min. After boiling, the suspension was snap-chilled at -20°C for 5 min. After chilling, cell debris was removed by centrifugation and 3 μl of the supernatant was used as a DNA template in PCR after quantitation and quality assessment of DNA using Nano-drop spectrophotometer. For identification of genus and species of Mycoplasma, the PCR was carried out in a final reaction volume of 25 µl using 200 µl capacity PCR tube containing 3 μl of DNA template, 1 μl of each primer (10 pmole/μl), 12.5 µl of 2X PCR Master-mix (Fermentas) and 7.5 μl of DNase-RNase free water. The isolates were confirmed as Mycoplasma sp. using genus specific forward, GPO-1 (5’-ACTCCTACGGGAGGCAGCAGTA-3’) and reverse, MGSO (5’-TGCACCATCTGTCACTCTGTTAACCTC-3’) primers amplifying 715 bp fragment of 16S rRNA of Mycoplasma sp. (Kuppeveld et al., 1992) after PCR. After initial denaturation at 94°C for 2 min, amplification was carried out for 30 cycles (consisting of denaturation at 94°C for 45 s, annealing at 55°C for 1 min and extension at 72°C for 1 min) with final extension at 72°C for 5 min. For confirmation of species as M. agalactiae, PCR was performed using M. agalactiae specific forward, Maga (5’-CCTTTTAGATTGGGATAGCGGATG-3’) and reverse Maga (5’-CCGTCAAGGTAGCGTCATTTCCTAC-3’) primers for an expected amplification product of 360 bp fragment of the 16S rRNA gene (Chávez-González et al., 1995). After initial denaturation at 95°C for 5 min, amplification was carried out for 40 cycles (consisting of denaturation at 94°C for 1 min, annealing at 57°C for 1 min and extension at 68°C for 1 min) with final extension at 70°C for 10 min. To detect the targeted amplification, 5 μl of PCR product from each tube was mixed with 1 μl of 6X gel loading buffer and electrophoresed on 1.5% agarose gel at a constant 80V for 30 min in 0.5X TBE buffer along with 100 bp DNA Ladder (GeneRuler-Fermentas). It was then stained with ethidium bromide (1% solution at the rate of 5 μl/100 ml), the size of the product was visualized under UV light and documented by the gel documentation system (SynGene, Gene genius bioImaging System, UK).
Amplification of p30 gene and restriction enzyme analysis
The membrane protein gene p30 of four representative isolates of M. agalactiae was amplified using the specific forward, P30(F) (5’-CAGGGGGATGAACATTTATG-3’) and reverse, P30(R) (5’- TTACCTCCATCTTTTTCAAC-3’) primers (Fleury et al., 2001) in a final reaction volume of 25 µl (containing 3 μl of DNA template, 1 μl of each primer (10 pmole/μl), 12.5 µl of 2X PCR master-mix (Fermentas) and 7.5 μl of DNase-RNase free water) for an expected amplicon of 730 bp. After initial denaturation for 2 min at 94°C, thermal cycling was carried out for 35 cycles (denaturation at 94°C for 30 s, annealing at 52°C for 45 s and extension for 1 min at 68°C) with final extension for 5 min at 72°C. The amplified PCR products of membrane protein gene were further processed and characterized by RE analysis. Four different REs, viz. RsaI, MboII, Sau3AI and AluI were selected from restriction map created using the sequences of p30 membrane protein gene available in GenBank at http://www.ncbi.nlm.nih.gov/Genbank/index.html and NEBcutter V2.0 software available online at http://tools.neb.com/NEBcutter2/index.php. A (30 µl) reaction mixture (containing 10 ml of PCR product, 1 ml of RE (10 U/µl), 2 μl of 10X restriction buffer and 17 μl of nuclease free water) was prepared and incubated in a water bath overnight according to the conditions specified by the manufacturer (Fermentas). After restriction digestion, an aliquot (10 ml) of each digested PCR product was mixed with 2 ml of gel loading buffer and electrophoresed along with 100bp DNA molecular weight marker on 2% agarose gel containing ethidium bromide (1% @ 5 ml/100 ml) by submarine gel electrophoresis apparatus at constant voltage of 60V for 45 min in 0.5X TBE buffer. After completion of electrophoresis, the gel was examined on UV transilluminator to observe the various fragments and photographed by gel documentation system (SynGene, Gene Genius BioImaging System,UK).
All the 13 isolates were identified on the basis of colony morphology and biochemical characters. In broth, floccular deposits were observed whereas on Modified Balanced Hank’s Salt Solution Agar (MBHS-A) medium, the typical fried egg appearance of colonies of Mycoplasmas were observed after staining. Biochemically, the isolates were sensitive to digitonin and positive for tetrazolium reduction, phosphatase production and film and spot test. All the isolates were negative for glucose metabolism, arginine hydrolysis and serum liquefaction. The isolates were further confirmed as M. agalactiae by PCR using genus and species specific primers which yielded specific amplification product of 715 bp and 360 bp respectively. The p30 gene with amplicon size of 730 bp was detected in all the representative M. agalactiae isolates analyzed (Figure 1). Digestion of the 730 bp product of the p30 gene with RsaI yielded a single large fragment of 654 bp along with the smaller 76 bp fragment which could not be resolved because of its smaller size (Figure 2). Analysis of the products with MboII revealed the presence of three fragments of 111 bp, 375 bp and 244 bp in size (Figure 3). Digestion of the amplified products with the Sau3AI yielded two fragments of 461 bp and 269 bp (Figure 4). Digestion of the amplified product with AluI yielded two fragments resolved into bands of 342 bp and 328 bp, whereas one smaller fragment of 60 bp could not be resolved (Figure 5).
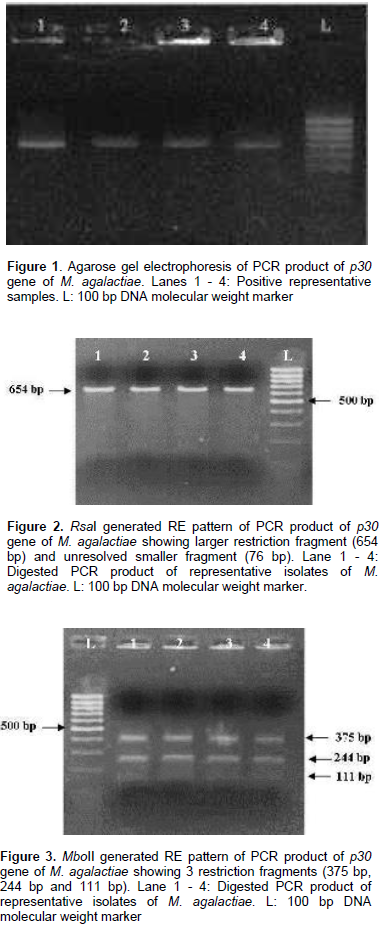
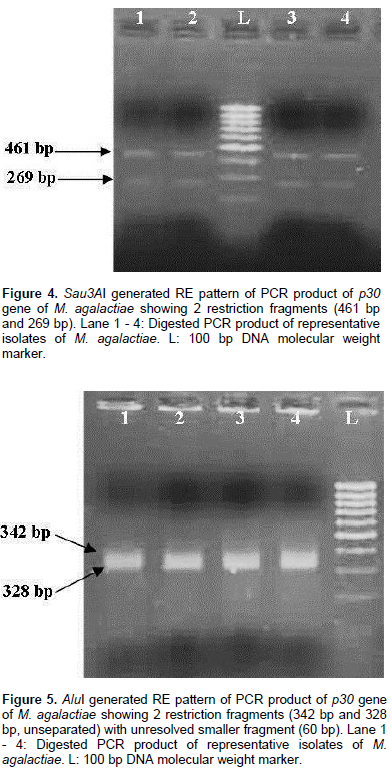
Restriction patterns produced by RsaI and MboII were in accordance with the expected restriction of M. agalactiae p30 gene revealing the presence of one and two restriction sites respectively. Variations in the expected pattern were observed in case of digestion with Sau3AI and AluI. Digestion with Sau3AI yielded only two fragments of 461 bp and 269 bp in contrast to the three expected fragments of 461 bp, 105 bp and 164 bp according to the restriction map showing the absence of one restriction site. Digestion of p30 amplicons with AluI revealed the absence of one restriction site resulting into a larger fragment of 328 bp in contrast to the expected two fragments of 282 bp and 46 bp along with other fragments of 60 bp and 342 bp fragments. Thus, the samples revealed the presence of only two restriction sites in contrast to the expected three restriction sites as per the expected restriction map with AluI. Several studies have been undertaken on the genetic characterization of the isolates by restriction digestion of various genomic segments of M. agalactiae including membrane protein gene. Glew et al. (2002) studied the polymorphism of the vpma locus of M. agalactiae by RE analysis using AseI, AlwI and HindIII. An identical banding pattern was observed for both AseI and AlwI whereas different patterns were observed with HindIII. In a similar study, Sung et al. (2006) analyzed the nested PCR products of various mycoplasmal strains by restriction enzyme digestion with Sau3AI enzyme to further identify and differentiate between the Mycoplasma species including M. agalactiae. Macun et al. (2010) targeted the 81 kDa membrane protein gene for the detection of M. agalactiae from cases of contagious agalactiae by PCR. In agreement with the present study, Kashoo et al. (2011) carried out the RE analysis of PCR products of p80 gene of M. agalactiae using RsaI and XhoI restriction enzymes to confirm the identity of the amplified products which produced the expected fragments of 146 and 868 bp with RsaI enzyme, while 176 and 838 bp fragments were obtained with XhoI enzyme on 2.5% agarose gel electrophoresis.
The present study reveals the presence of polymorphism using PCR-RE analysis among the strains of M. agalactiae prevailing in the field in Gujarat region of India. This western province has significant population of sheep and goat which are affected by several disease condition and many disease outbreaks have been reported in them. Screening of those flocks is important to understand the cause of morbidity and mortality. Molecular tools are very important for rapid screening of those flocks as well as for characterization of the associated pathogens. The present study also shows the importance of molecular tools in the study of genomic variation among the field strains of these microorganisms causing covert and overt manifestations utilizing various mechanisms to evade host immune response. Many studies globally have shown the importance of RE analysis in the study of polymorphism in various genomic segments of field strains of M. agalactiae and development of detection system for different species. These findings further underline the need to study the immunological implications of these variations occurring in the membrane proteins of these mycoplasmal pathogens which can ultimately affect their pathogenesis and persistence in the host.
The authors have not declared any conflict of interests.
The authors are thankful to the Professor and Head, Department of Veterinary Microbiology and Dean, College of Veterinary Science and Animal Husbandry, Anand (Gujarat), for providing the facilities to pursue this work.
REFERENCES
|
Cacciotto C, Addis MF, Pagnozzi D, Chessa B, Coradduzza E, Carcangiu L, Uzzau S, Alberti A, Pittau M (2010). The liposoluble proteome of Mycoplasma agalactiae: an insight into the minimal protein complement of a bacterial membrane. BMC Microbiol. 10:225.
Crossref
|
|
|
|
Carmichael LE, Sullivan ND, Hoasfall N (1972). Isolation, propagation and characterization studies of an ovine Mycoplasma responsible for proliferative interstitial pneumonia. Cornell. Vet. 62:654-679.
|
|
|
|
Chávez-González Y, Bascunana CR, Bolske JG, Mattson JB, Molina CF, Johansson KE (1995). In vitro amplification of the 16S rRNA genes from Mycoplasma bovis and Mycoplasma agalactiae by PCR. Vet. Microbiol. 47:183-190.
Crossref
|
|
|
|
Chopra-Dewasthaly R, Spergser J, Zimmermann M, Citti C, Jechlinger W, Rosengarten R (2017). Vpma phase variation is important for survival and persistence of Mycoplasma agalactiae in the immunocompetent host. PLoS Pathog. 13(9):e1006656.
Crossref
|
|
|
|
de Garnica ML, Rosales RS, Gonzalo C, Santos JA, Nicholas RAJ (2013). Isolation, molecular characterization and antimicrobial susceptibilities of isolates of Mycoplasma agalactiae from bulk tank milk in an endemic area of Spain. J. Appl. Microbiol. 114:1575-1581.
Crossref
|
|
|
|
Erno H, Stipkovits L (1973). Bovine Mycoplasmas: cultural and biochemical studies. Acta. Vet. Scand. 14:436-449.
|
|
|
|
Fleury B, Bergonier D, Berthelot X, Peterhans E, Frey J, Vilei EM (2002). Characterization of P40, a cytadhesin of Mycoplasma agalactiae. Infect. Immun. 70(10):5612-21.
Crossref
|
|
|
|
Fleury B, Bergonier D, Berthelot X, Schlatter Y, Frey J, Vilei EM (2001). Characterization and analysis of a stable serotype-associated membrane protein (p30) of Mycoplasma agalactiae. J. Clin. Microbiol. 39(8):2814-2822.
Crossref
|
|
|
|
Flitman-Tene R, Levisohn S, Lysnyansky I, Rapoport E, Yogev D (2000). A chromosomal region of Mycoplasma agalactiae containing vsp-related genes undergoes in vivo rearrangement in naturally infected animals. FEMS Microbiol. Lett. 191:205-212.
Crossref
|
|
|
|
Freundt EA, Ernø H, Black FT, Krogsgaard-Jensen A, Rosendal S (1973). Evaluation of reference reagents for Mycoplasmas. Ann. N. Y. Acad. Sci. 225:161-171.
Crossref
|
|
|
|
Glew MD, Marenda M, Rosengarten R, Citti C (2002). Surface diversity in Mycoplasma agalactiae is driven by Site-Specific DNA inversions within the vpma Multigene Locus. J. Bact. 184(21):5987-5998.
Crossref
|
|
|
|
Kashoo ZA, Singh VP, Rana R, Sankar M, Gazalli H, Susan S (2011). Molecular characterization of P80 gene of Indian Mycoplasma agalactiae isolates. Ind. J. Anim. Sci. Sci. 81(5):3-7.
|
|
|
|
Macun HC, Ocal N, Karahan M, Yagci BB, Kalender H, Kalim, R (2010). Endemic contagious agalactiae in sheep and goats: Clinical evaluation, Treatment and Vaccination. J. Anim. Vet. Adv. 9(14):1918-1924.
Crossref
|
|
|
|
Madanat A, Zendulkova D, Pospisil Z (2001). Contagious agalactia of sheep and goats: A review. Acta Vet. Brno. 70:403-412.
Crossref
|
|
|
|
Razin S, Yogev D, Naot Y (1998). Molecular biology and pathogenicity of mycoplasmas. Microbiol. Mol. Biol. Rev. 62:1094-1156.
|
|
|
|
Solsona M, Lambert M, Poumarat F (1996). Genomic, protein homogeneity and antigenic variability of Mycoplasma agalactiae. Vet. Microbiol. 50(1-2):45-58.
Crossref
|
|
|
|
Sung H, Kang SH, Bae YJ, Hong JT, Chung YB, Lee C, Song S (2006). PCR based detection of Mycoplasma species. J. Microbiol. 44(1):42-49.
|
|
|
|
Van-Kuppeveld FJ, Van-der-logt JT, Angulo AF, Van-Zoest MJ, Quint WG, Niesters HG, Galama JM, Melchers WJ (1992). Genus and species specific identification of Mycoplasmas by 16S rRNA amplification. Appl Environ Microbiol. 58:2606-2615.
|