ABSTRACT
Silver nanoparticles (AgNPs) have stimulated interest of scientists due to their wide range of applications, including their potential antimicrobial activity. This study investigated the antimicrobial activities of AgNPs synthesized by rhizospheric soil and fish pond sediment microorganisms against selected clinical pathogens. The samples were cultured and organisms identified in accordance with standard procedures. The synthesis of AgNPs colloidal solution was monitored by UV-vis analysis. Presence of bands was determined by the Fourier Transform Infrared spectroscopy (FTIR). The antimicrobial activity of the synthesized AgNPs against selected clinical isolates was determined using agar well diffusion method. Ten species each of bacteria and fungi were isolated from the samples. Formation of AgNPs was indicated by colour transformation from yellow to brown. All synthesized AgNPs showed intense peak with wavelengths ranging of 410-440 nm in UV-vis. The FTIR revealed band at 3395 cm-1 and a strong peak at 3300-3500 cm-1. The AgNPs synthesized by some of the isolates exerted remarkable and varying degrees of antimicrobial activities against the susceptible test organisms. This study revealed that the microbially-synthesized AgNPs obtained from this study possess a high antimicrobial potency against most potential pathogens investigated, and, thus, can be exploited in the development of novel antimicrobial agents.
Key words: Antimicrobial agents, microbial resistance, microorganisms, pathogens, silver nanoparticles, zones of inhibition.
Multidrug resistance is a major concern in the treatment of infectious diseases. The wide and indiscriminate use of broad-spectrum antibiotics has led to resistance to traditional antimicrobial agents for many bacterial human pathogens, and this is a major threat to the global health care (Amr-Saeb et al., 2014). Hence, a new research for potent antimicrobial agent is necessary. Nanotechnology is a relatively new field applied in research, especially in biotechnology. New applications of nanoparticles and nanomaterials are also increasing rapidly. Biological method of synthesis provides a wide range of environmentally acceptable methodology with low cost of production and minimum time required (Kannan et al., 2010; Al-Khuzai et al., 2019) since the reducing agent and the stabilizer used during chemical synthesis are replaced by molecules obtained from living organisms such as bacteria, fungi, yeasts, algae, or plants (Narayanan and Sakthivel, 2010). The unique properties of AgNPs such as the size, shape, electrical, and magnetic properties, can be incorporated into antimicrobial applications, biosensor materials, composite fibers, cryogenic superconducting materials, cosmetic products, and electronic components, and these have stimulated researchers’ interests in their applications in these fields (Vishwanatha et al., 2018; El-Saadony et al., 2019). Several methods have been used for synthesizing and stabilizing AgNPs such as laser ablation, chemical reduction, gamma irradiation, electron irradiation, microwave processing, photochemical and biological synthetic methods, (Ghareib et al., 2016; Vishwanatha et al., 2018; El-Saadony et al., 2019).
However, the synthesis of AgNPs from bacteria cells has gained much attention because bacterial cells possess a special mechanism of resistance to silver ions in the environment. This innate feature is responsible for growth and survival in the environments with metal ion concentrations and their ability to synthesize nanoparticles (Saklani et al., 2012). Efflux systems, alteration of solubility and toxicity which occur via extracellular complex formation or precipitation of metals, biosorption, bioaccumulation, reduction or oxidation, and lack of specific metal transport systems are the mechanisms involved in resistance (Husseiny et al., 2006).
Silver, when compared with other metals, shows stronger toxicity to microorganisms and the advancement in the field of nanotechnology has tremendously assisted researchers to explore novel means of developing more potent antimicrobial drugs. As a result of the fact that silver and its compounds possess high antimicrobial activity, silver nanoparticles (AgNPs) have stimulated interest of scientists owing to their wide range of applications (Qais et al., 2019). The prevalence of infectious diseases is a worldwide challenge and the problem of development antimicrobial resistance continues to become a global health risk. This necessitates the investigation on the biosynthesis of silver nanoparticles from microorganisms from environmental samples with the determination of their antibacterial and antifungal potentials (Keshavamurthy et al., 2017).
It is widely accepted that the presence of nitrate reductase is solely responsible for biosynthesis of AgNPs. Nitrate reductase is responsible for the conversion of nitrate to nitrite (Keshavamurthy et al., 2017). The in vitro demonstration of this mechanism was reported by Kalimuthu et al. (2008) using Bacillus licheniformis. The bacterium secreted cofactor NADH and NADH-dependent enzymes, especially nitrate reductase which were speculated to be responsible for the bioreduction of silver ion (Ag+) to Ag0 and the subsequent formation of AgNPs. Anil-Kumar et al., (2007) confirmed this speculation with the first direct evidence for the involvement of nitrate reductase in the synthesis of AgNPs.
Fungi have the ability of reducing the metals ions into their corresponding nanometals either intracellularly or extracellularly due to their high binding capacity with metal (Al-Khuzai et al., 2019). They were found to produce larger amounts of nanoparticles compared to bacteria because they can secrete larger amounts of proteins which directly translate to higher productivity of nanoparticles (Mohanpuria et al., 2008). Fungi are unique and can be considered as the best producers of nanoparticlees in relation to bacteria because they are easy to culture on solid substrate fermentation and they can grow on the surface of inorganic substrate during culture leading to efficient distribution of metals as catalyst (Ahmad et al., 2003). Extracellular production of nanoparticles from fungi also has an advantage of producing large quantity of enzymes which are in pure state and free from cellular protein, and which are easy to apply for the simple downstream process. Therefore, this study investigated the biosynthesis and antimicrobial potentials of AgNPs obtained from microorganisms associated with rhizospheric soil and fish pond sediments from the Federal University of Technology, Akure, FUTA, Nigeria.
Collection of samples
Samples of rhizospheric soil of selected plants (guava, banana, mango, and cassava) and fish pond sediment were collected at different areas and varying depth in FUTA. The samples were taken from a depth of 5-10 cm on the farm then kept in plastic bags and transported immediately to the laboratory for analyses.
Collection of test organisms
Five clinical bacterial pathogens -Pseudomonas aeruginosa, Escherichia coli, Klebsiella pneumoniae, Staphylococcus aureus and Enterococcus faecalis–and five fungal pathogens – Aspergillus fumigatus, Penicillium notatum, Rhizopus stolonifer, Aspergillus flavus and Trichoderma viride - were collected from Ondo State Specialist Hospital, Akure, Nigeria, and resuscitated on nutrient agar media to obtain young actively growing culture.
Microbiological analysis of samples
Rhizospheric soil and fish pond sediment samples were separately sieved with 0.5 mm sieve to remove stones and plant debris. Thereafter, 1 g of each sample was serially diluted to obtain a six-fold dilution factor. An aliquot (0.1 ml) of each dilution was aseptically inoculated on appropriate media. The plate count agar, cetrimide agar, mannitol salt agar, eosin methylene blue agar and blood agar media (Oxoid, England) were used for the cultivation of total heterotrophic bacteria, P. aeruginosa, S. aureus, E. coli and Bacillus species, respectively using the pour plate technique. This method was also adopted for cultivation of fungi on potato dextrose agar medium. All bacterial culture plates were incubated at 32°C for 24 h while fungal were incubated at 28°C for 72 h. Pure cultures were obtained by repeated subcultures on fresh media using the streak plate technique (Harrigan and McCane, 1976; Bello et al., 2012).
Identification of microorganisms
Primary identification was done by observation of cultural characteristics of pure isolates while characterization procedures were carried out as described by Cowan and Steel (1985) and Holt et al. (2004). Gram’s staining reactions and cell morphology from heat-fixed smears were observed. The motility of isolates was determined by the hanging drop technique, and biochemical tests were carried out on the isolates in accordance with standard procedures. Fungal isolates were identified as described by Beneke and Rogers (2005). The Analytical Profile Index (API) 20E and API 20NE were used for the additional identification of the families Enterobacteriaceae and non-Enterobacteriaceae, respectively. API 20C was used for further identification of fungal isolates. These were done in accordance with the manufacturers’ protocols (BioMerieux, Marcy I’Etoile, France).
Synthesis and characterization of AgNPs from microbial isolates
The bacteria isolated from the rhizospheric soil were cultured on both nutrient broth as well as Luria Bertani broth to produce biomass for biosynthesis. The pH was adjusted to 7.0. Incubation was done on an orbital shaker at 27°C at 220 rpm. The biomass of each isolate was harvested after 24 h and centrifuged at 12000 rpm for 10 min. The supernatant was collected and used for extracellular synthesis of AgNPs. The supernatant was added separately to the reaction vessel containing 1 mM of silver nitrate (AgNO3) while the control was set up without the silver nitrate for 24 h in the dark. The reduction of the Ag+ ions in the solution was monitored by observing changes of the colour. The absorbance was measured at a resolution of 1 nm using UV–visible spectrophotometer with samples in quartz cuvette (Kannan et al., 2010; Vanmathi and Sivakumar, 2012).
The chemical compounds of the synthesized AgNPs were studied by using Fourier Transformed Infra-Red Spectrophotometer (FTIR) (Perkin-Elmer LS-55-Luminescence spectrometer). The solutions were dried at 75°C and the dried powders were characterized to identify possible interactions between the Ag salts and protein molecules which could account for the reduction of silver ions and the stabilization of AgNPs (Vanmathi and Sivakumar, 2012).
Transmission electron microscopy (TEM) analysis of AgNPs
In order to obtain quantitative measures of AgNPs, their size distribution and morphology, analysis of the samples was performed using TEM technique. The size and shape distributions of produced AgNPs were characterized by adding a few drops of AgNPs solution onto a TEM grid, and the residue was removed by a filter paper beneath the TEM grid (Mohammadi et al., 2019). The magnification of TEM was determined by the ratio of the distance between the objective lens and the specimen, and the distance between objective lens and its image plane.
Antimicrobial activities of the microbially-synthesized AgNPs against clinical isolates
Antimicrobial activities of AgNPs synthesized by microorganisms from the rhizospheric soil and fish pond sediments were tested against P. aeruginosa, E. coli, K. pneumoniae, S. aureus, E. faecalis, A. fumigatus, P. notatum, R. stolonifer, A. flavus and T. viride using the agar well diffusion assay method as described by Ozcelika et al. (2006). A 20 µl portion of the nanoparticles solution was taken and introduced on Mueller Hinton agar plate incubated at 37°C for 24 h. Ciprofloxacin and ketoconazole were used as positive control for bacteria and fungi, respectively. Zones of inhibition exerted by the microbially-nanoparticles were measured in millimeters with the aid of a metre rule.
Identification of microorganisms isolated from rhizospheric soil and fish pond sediment
The ten bacterial species isolated from the rhizospheres and fish pond sediments were characterized as S. aureus, S. epidermidis, P. vulgaris, E. coli, Micrococcus luteus, P. aeruginosa, Bacillus subtilis, B. cereus, Serratia marcescens and Streptomyces griseus (Appendix Table 1). Ten fungal species were also identified and these include Penicillium frequentans, Saccharomyces cerevisiae, Aspergillus niger, A. flavus, Fusarium oxysporum, R. stolonifer, Mucor mucedo, Emericella rugulosa, T. viride and Geotrichum albidum (Appendix Table 2).
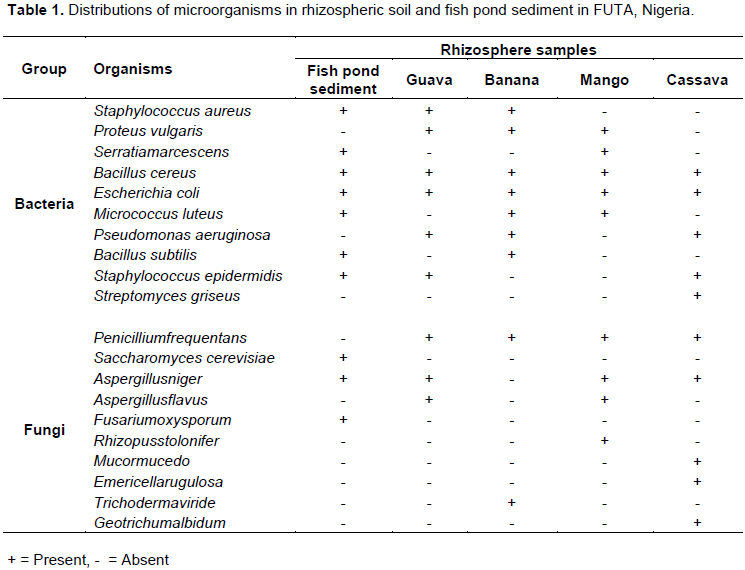
Distributions of microorganisms in the rhizospheric soil and fish pond sediment
S. aureus was isolated from the fish pond sediment sample and rhizospheric soils of guava and banana plants. B. cereus and E. coli were encountered in the fish pond sediment and rhizospheric soil samples of guava, banana, mango and cassava plants investigated in this study. P. vulgaris was present in the rhizospheric soils of guava, banana and mango. S. marcescens was encountered only in the rhizospheric soil of mango and the fish pond sediment while B. subtilis was found in the fish pond sediment and the rhizospheric soil of banana only. S. cerevisiae, A. niger and F. oxysporum were present in the fish pond sediment. P. freqentens, A. niger and A. flavuswere found in the rhizospheric soils of guava and mango plants in addition to R. stolonifer encountered in only mango plants. Only P. freqentens and T. viride were encountered in the soil associated with banana plant while P. freqentens, A. niger, M. mucedo, E. rugulosa and G. albidum were present in rhizospheric soil of cassava plants (Table 1).
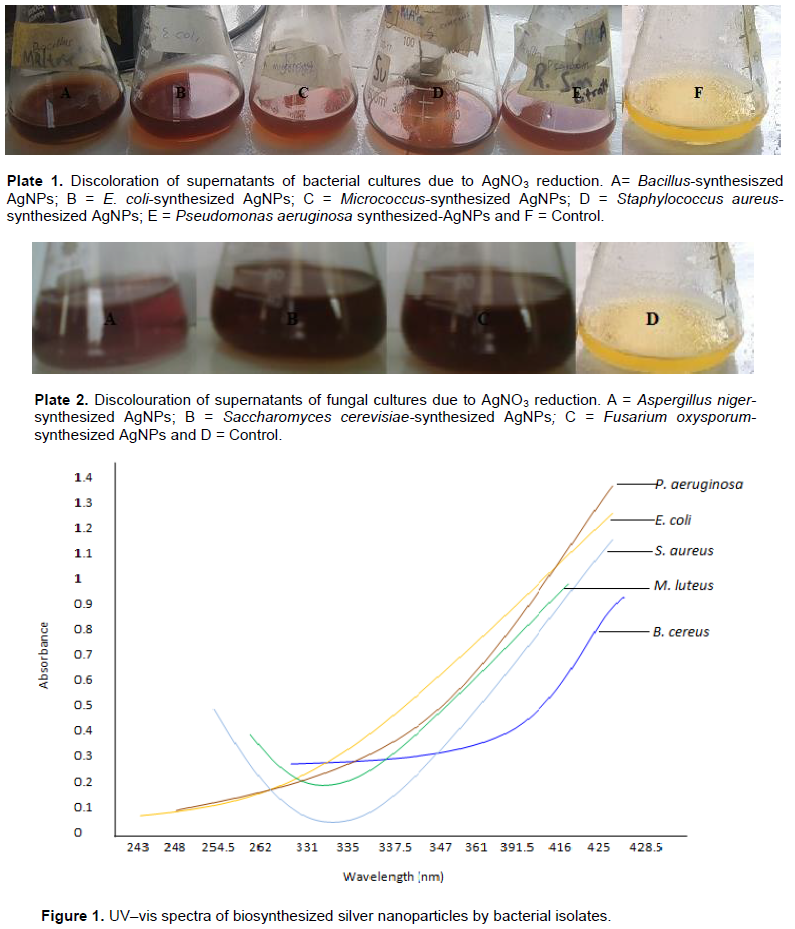
Colour change and UV-vis analysis
The appearance of a yellowish-brown colour in the silver nitrate treated flask indicated the formation of AgNPs (Plates 1 and 2). Among the bacterial species isolated in this study, B. subtilis, E. coli, M. luteus, P. aeruginosa and S. aureus (Figure 1) were found to synthesize AgNPs with intense peak in wavelength ranging from 416-428 nm while the fungi, which included Aspergillus niger, S. cerevisiae and Fusarium oxysporum (Figure 2), showed broad and strong spectra ranging from 417-437.5 nm.
FTIR analysis of silver nanoparticles
The FTIR analysis was used to identify possible interactions between the Ag salts and protein molecules in the reduction of silver ions and the stabilization of AgNPs. The bacterial and fungal isolates revealed the existence of spectral band at 3395 cm-1 in the FTIR spectrograms and a strong peak at 3300-3500 cm-1. The peak at 2185 cm-1 was also significant to most of the isolates. Peaks were also observed at 1675 cm‑1 in all the isolates as shown in Figure 3A to E for bacterial species and Figure 4A to C for fungal species.
Transmission electron microscopy (TEM) analysis
To gain further insight into the features of the AgNPs, analysis of the sample was performed using TEM technique. The shape and size of the obtained AgNPs were elucidated with the aid of TEM images. Nanoparticles observed from the micrographs were majorly spherical with a small percentage of elongated particles ranging in size from 5 to 30 nm with an average size of 20 nm (Figure 5).
Antimicrobial potency of AgNPs synthesized by the microbial species against selected clinical organisms
The B. subtilis-synthesized AgNPs exerted the highest potency against the test organisms as compared with other AgNPs synthesized by other Gram-positive isolates in the study. B. subtilis-synthesized AgNPs showed antibacterial activities against S. aureus, E. coli, and E. faecalis with zones of inhibition of 5.33, 7.00 and 14.33 mm, respectively. The antimicrobial activity of AgNPs synthesized by B. subtilis against E. faecalis was even higher than that exerted by ciprofloxacin (the control) with zone of inhibition of 10.67 mm (Table 2).
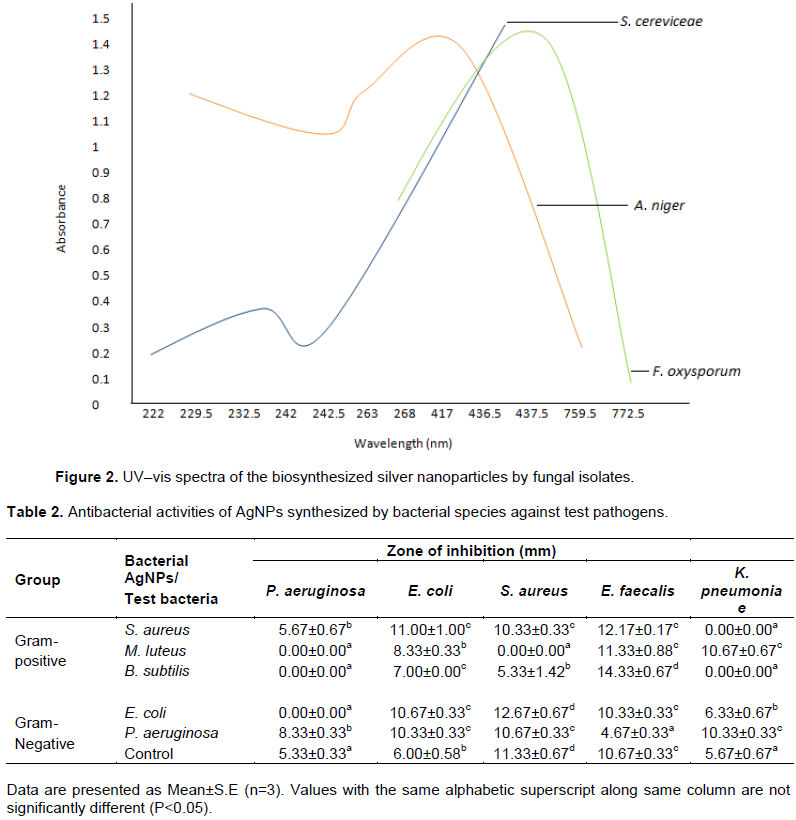
E. coli-synthesized AgNPs exhibited antibacterial activity against K. pneumoniae, E. faecalis, E. coli and S. aureus with zones of inhibition of 6.33, 10.33, 10.67 and 12.67 mm, respectively. There was no significant difference between this antibacterial effect on E. coli and E. faecalis (P > 0.05). P. aeruginosa-synthesized AgNPs also exerted varying degrees of antibacterial activity against all the test pathogens with zones of inhibition ranging from 4.67 to 10.67 mm. The activities on E. coli, K. pneumoniae and S. aureus showed no statistical difference (P > 0.05) (Table 2).
The antifungal activity of AgNPs synthesized by three of the fungal isolates against selected fungal pathogens is shown in Table 3. The A. niger-synthesized AgNPs exerted antifungal effect on R. stolonifer, P. notatum, A. flavus and T. viride with inhibition zones of 2.33, 5.33, 10.67 and 10.67 mm, respectively. There was no statistical difference between the antifungal activity exerted against A. flavus and T. viride (P > 0.05) while that of P. notatum and R. stolonifer showed significant difference (P < 0.05). The AgNPs synthesized by S. cerevisiae exerted the highest antimicrobial potency against A. fumigatus and P. notatumas compared with other fungal species in that category, and even the control antibiotic (ketoconazole).
The distribution of microorganisms encountered in this study varied and each sample contributed to the microbial diversity investigated. Wolfe and Kilironomas (2005) reported that the quality of root exudates can promote differential recruitment of microorganisms present in the soil. The presence of nitrogen-fixing bacteria in the rhizosphere of plants can also improve plant growth in nitrogen-poor environments, as well as promote increased nitrogen content in the soil, which is often related to the facilitative effect that legume species have on other plant species (Walker, 2003).
The colour transformation from yellow to brownish indicates the formation of AgNPs (Singh et al., 2011). The intensity of the brown colour increased dramatically up to 24 h and this may be as a result of the excitation of surface plasmon resonance (SPR) and the reduction of AgNO3 (Manivasagan et al., 2013; Deljou and Goudarzi, 2016). AgNPs mostly had an absorption peak at 425 nm attributed to their SPR probably due to the stimulation of longitudinal plasmon vibrations (Fig. 1) and which is in line with the report of Kumar and Mamidyala (2012). The SPR property is also responsible for the colour change of the reaction mixture from yellowish to brown (Chaudhari et al., 2012; Yamal et al., 2013). Therefore, the increase of the absorbance at 347 nm is a reliable criterion indicating AgNPs synthesis (Thu et al., 2013).
The FTIR analysis is used for the detection of potential interactions between silver salt that are involved in AgNPs formation (Saha et al., 2010), while it has been suggested that this analysis provides information on the binding of proteins to AgNPs which leads to nanoparticles stabilization (Jain et al., 2011). For these reasons, FTIR has been routinely used by several researchers in nanoparticles characterization (Deljou and Goudarzi, 2016). Similar FTIR spectra were obtained by microbial isolates. They all indicated the existence of proteins in the capping agent of the nanoparticles, and also that the secondary structure of proteins was not affected as a consequence of the reaction with silver ions or the binding to AgNPs. This assertion is supported by the band at 3395 cm−1 in the FTIR spectrogram, which is specific to the frequency of extending vibration of primary amines, and the strong band peak at 3300–3500 cm−1, which characterizes the stretching vibrations of N-H, indicating strong hydrogen bonding. This is in accordance with the findings of Saha et al. (2010), who reported 3300-3500 cm-1 as strong band peak.
The appearance of a band at about 2185 cm−1, which is assigned to C=O extend vibrations of carboxylic acids, aldehydes and ketones, was remarkable, indicating that the oxidation of the hydroxyl groups of hydrolysates, which is known to originate from the medium peptides, are associated with the reduction of silver ions. The bands observed at 1675 cm−1 is a definite indicator of linkages between the amides I and II (Sharma et al., 2012). It is well known that proteins can bind to the AgNPs either through free amine groups or cysteine residues in the proteins (Gole et al., 2001). FTIR spectroscopy thus revealed the possible stabilization of AgNPs with proteins (Venkatesan et al., 2013).
Presence of spectral peaks at 3300 - 3500 cm−1 may be attributed to aliphatic C-H stretching vibration of hydrocarbon chains and N-H bending vibration (Cheng et al., 2014). The spectral vibration of aldehydic group (C=O) was shown by wave length 3395 cm−1 (Hamouda et al., 2019) while the peak at the range 1675 cm−1 could be attributed to amides (N-H) stretching in addition to peptide bond and C=C stretching involved in stabilizing nanoparticles by proteins as explained by Castro et al. (2013). A spectral band at 3395 cm−1 could be designated to the residual amount of AgNO3 (ŠedÄ›nková et al., 2009). The presence of absorption bands at 1675 cm−1 may also be attributed to vibration of the -C-O group (Hamouda et al., 2019). Spectral peaks at 242.5-417, 242-436.5 and 437.5 cm−1 indicated the bending region of the aliphatic chain. Moreover, bands assigned at 3395 cm−1 could be attributed to either phosphorus or sulfur functional groups, which possibly attach silver and perform both capping and stabilizing process of nanoparticles (Castro et al., 2013). It had been reported that biologically-synthesized AgNPs are promising therapeutic agents with significant antimicrobial activities (Galdiero et al., 2011; Mohammadi et al., 2019), and a number of these biosynthesized nanoparticles had been described and characterized based on their ability to inhibit microbes (Deljou and Goudarzi, 2016). Vishwanatha et al. (2018) also reported that the biogenic and eco-friendly route for synthesizing AgNPs with antibacterial activity against clinically important pathogens and attributes growing interest on fungi as an emerging source for the synthesis of NPs.
The antibacterial activity of AgNPs synthesized by Gram-positive bacteria revealed that AgNPs from B. subtilis showed inhibition on Enterococcus faecalis, Escherichia coli, and Staphylococcus aureus respectively. The result of this research is in accordance with the findings of Deljou and Goudarzi (2015) who reported that AgNPs synthesized by Bacillus spp. showed significant inhibition on some selected human pathogens. The AgNPs synthesized from S. aureus showed significant inhibition on all the selected isolates except K. pneumoniae. The AgNPs synthesized by M. luteus showed significant inhibition on E. faecalis, and K. pneumoniae. This research revealed that only S. aureus-synthesized AgNPs showed inhibition on Pseudomonas aeruginosa.
Antibacterial activity of Gram-negative-bacteria-synthesized AgNPs revealed that P. aeruginosa-synthesized AgNPs showed inhibition on all the selected isolates while E. coli-synthesized AgNPs showed inhibition on all the selected isolates except P. aeruginosa. The efficacy of AgNPs can be attributed to the fact that their larger surface area gives them a better contact with the microorganisms. This is further supported by the revelation that size dependent interaction of AgNPs with bacteria leads to its antibacterial activity (Pal et al., 2007). The nanoparticles get attached to the cell membrane and also penetrate inside the bacterial cells (Venkatesan et al., 2013). The bacterial membrane is known for its sulphur containing proteins, these might be the preferential sites for the AgNPs to penetrate (Venkatesan et al., 2013). The A. niger-synthesized AgNPs exerted antifungal effect on R. stolonifer, P. notatum, A. flavus and T. viride while it had activity against A. fumigatus. The F. oxysporum-synthesized AgNPs exhibited antifungal activities against T. viride, A. fumigatus, P. notatum and A. flavus but no activity was exerted against R. stolonifer. The S. cerevisiae-synthesized AgNPs exhibited activity against R. stolonifer, A. fumigatus and P. notatum while no activity was shown against A. flavus and T. viride.
The mode of action of AgNPs is triggered by the generation of reactive oxygen species (ROS) inside both bacterial and fungal cells. Feng et al. (2008) reported that nanoparticles are capable of releasing silver ions, while Matsumura et al. (2003) added that these ions can interact with the thiol groups of many vital enzymes and inactivate them. Silver ion is taken in by microbial cells that come in contact with silver which leads to inhibition of several functions in the cell, and thus, damage the cells. The inhibition of a respiratory enzyme by silver ion leads to the generation of reactive oxygen species, which attacks the cell itself (Prabhu and Poulose, 2012).
On the other hand, the toxicity of silver ions, could be by their adhesion to the cell membrane and further penetration inside or by interaction with phosphorus containing compounds like DNA disturbing the replication process or preferably by their attack on the respiratory chain. It has also been suggested that a strong reaction takes place between the silver ions and thiol groups of vital enzymes thus inactivating them (Sunkar and Nachiyar, 2012; Venkatesan et al., 2013). Also, the changes in morphology of bacterial membrane as well as the possible damage caused by the nanoparticles reacting with the DNA will affect the bacteria in cell processes such as the respiratory chain and cell division, finally causing cell death (Sondi and Salopek-Sondi, 2004). Furthermore, nanoparticles might also release silver ions in the bacterial cells, which further enhance their bactericidal activity (Morones et al., 2005).
The shapes and sizes of the obtained AgNPs produced by the bacterial and fungal species were projected by TEM images. The structures of the AgNPs produced by the species of bacteria were identical, which could be attributed to a similarity in the reductive agents present in the species (Jyoti et al., 2016). The AgNPs produced by the bacterial species were well-dispersed without any visible agglomeration or morphological variations. The observations in relation to shapes, sizes and dispersion were same for the AgNPs produced by fungal species. However, the fungi-mediated biosynthesized AgNPs were better dispersed than those of the bacterial species. These reports are in agreement with Singh et al., (2017) and Abdel-Raouf et al. (2018).
This study revealed that microbially-synthesized AgNPs possess a high antimicrobial potency against most pathogens, and, thus, can be exploited in the development of novel antimicrobial agent.
The authors have not declared any conflict of interests.
REFERENCES
|
Abdel-Raouf N, Al-Enazi NM, Ibraheem IBM, Alharbi RM, Alkhulaif MM (2018). Biosynthesis of silver nanoparticles by using of the marine brown alga Padina pavonia and their characterization. Saudi Journal of Biological Sciences 26(6):1207-1215.
Crossref
|
|
|
|
Ahmad A, Mukherjee P, Senapati S, Mandal D, Khan MI, Kumar R, Sastry M (2003). Extracellular biosynthesis of silver nanoparticles using the fungus Fusarium oxysporum. Colloids and Surfaces B: Biointerfaces 28:313-318.
Crossref
|
|
|
|
|
Al-Khuzai RAH, Aboud MK, Alwan SK (2019). Biological Synthesis of Silver Nanoparticles from Saprolegnia parasitica. Journal of Physics: Conference 1294(6):1-15.
Crossref
|
|
|
|
|
Amr-Saeb TM, Alshammari AS, Al-Brahim H, Al-Rubeaan KA (2014). Production of Silver Nanoparticles with Strong and Stable Antimicrobial Activity against Highly Pathogenic and Multidrug Resistant Bacteria. The Scientific World Journal.
Crossref
|
|
|
|
|
Anil-Kumar S, Abyaneh MK, GosaviSulabha SW, Ahmad A, Khan MI (2007). Nitrate reductase mediated synthesis of silver nanoparticles from AgNO3. Biotechnology Letters 29:439-445.
Crossref
|
|
|
|
|
Bello OO, Mabekoje OO, Efuntoye MO, Bello TK (2012). Prevalence of vaginal pathogens associated with genital tract infections in Ogun State, Nigeria. British Microbiology Research Journal 2(4):277-289.
Crossref
|
|
|
|
|
Beneke ES, Rogers AL (2005). Medical mycology manual. 3rd ed. Minneapolis: Burgers Publishing P 226.
|
|
|
|
|
Castro L, Blázquez ML, Muñoz JA, González F, Ballester A (2013). Biological synthesis of metallic nanoparticles using algae. IET Nanobiotechnology 7(3):109-116.
Crossref
|
|
|
|
|
Chaudhari PR, Shalaka AM, Vrishali BS, Suresh PK (2012). Antimicrobial activity of extracellular synthesized silver nanoparticles using Lactobacillus species obtained from VIZYLAC capsule. Journal of Applied Pharmaceutical Science 2(3):25-29.
|
|
|
|
|
Cheng KM, Hung YW, Chen CC, Liu CC, Young JJ (2014). Green synthesis of chondroitin sulfate-capped silver nanoparticles: Characterization and surface modifcation. Carbohydrate Polymers 110:195-202.
Crossref
|
|
|
|
|
Cowan ST, Steel KJ (1985). Manual for the identification of bacteria. Cambridge University Press, Verlage, New York, P. 502.
|
|
|
|
|
Deljou A, Goudarzi S (2016). Green extracellular synthesis of the silver nanoparticles using thermophilic Bacillus sp. AZ1 and its antimicrobial activity against several human pathogenic bacteria. Iranian Journal of Biotechnology 14(2):25-32.
Crossref
|
|
|
|
|
El-Saadony MT, El-Wafai NA, El-Fattah HIA, Mahgoub SA (2019). Biosynthesis, optimization and characterization of silver nanoparticles using a soil isolate of Bacillus pseudomycoides MT32 and their antifungal activity against some pathogenic fungi. Advances in Animal and Veterinary Sciences 7(4):238-249.
Crossref
|
|
|
|
|
Feng QL, Wu J, Chen GQ, Cui FZ, Kim TN, Kim JO (2008). A mechanistic of the antibacterial effect of silver ions on E. coli and S. aureus. Journal of Biomedical Materials Research 52(4):662-668.
Crossref
|
|
|
|
|
Galdiero S, Falanga A, Vitiello M, Cantisani M, Marra V, Galdiero M (2011). Silver nanoparticles as potential antiviral agents. Molecules 16(10):8894-8918.
Crossref
|
|
|
|
|
Ghareib M, Tahon MA, Saif MM, El-Sayed AW (2016). Rapid Extracellular Biosynthesis of Silver Nanoparticles by Cunning hamellaphaeospora Culture Supernatant. Iranian Journal of Pharmaceutical Research 15(4):915-924.
|
|
|
|
|
Gole A, Dash C, Ramakrishnan V, Sainkar V, Mandale AB, Rao M (2001). Pepsin-Gold Colloid Conjugates: Preparation, Characterization, and Enzymatic Activity. Langmuir 17(5):1674-1679.
Crossref
|
|
|
|
|
Hamouda RA, Hussein MH, Abo-elmagd RA, Bawazir SS (2019). Synthesis and biological characterization of silver nanoparticles derived from the cyanobacterium Oscillatoria limnetica. Scientific Report 9:13701.
Crossref
|
|
|
|
|
Harrigan MG, McCane ME (1976). Laboratory methods in food and dairy microbiology. London: Academic Press pp. 33-200.
|
|
|
|
|
Holt GH, Krieg NR, Sneath PH, Staley JT, Williams ST (2004). Bergey's manual of determinative bacteriology. 9th ed. Baltimore: Williams and Wilkins, 787 p.
|
|
|
|
|
Husseiny M, Aziz MAE, Badr Y, Mahmoud MA (2006). Biosynthesis of goldnanoparticles using Pseudomonas aeruginosa. Spectrochimica Acta Part A: Molecular Spectroscopy 67(3-4):1003-1006.
Crossref
|
|
|
|
|
Jain N, Bhargava A, Majumdar S, Tarafdar JC, Panwar J (2011). Extracellular biosynthesis and characterization of silver nanoparticles using Aspergillus flavus NJP08: a mechanism perspective. Nanoscale 3(2):635-41.
Crossref
|
|
|
|
|
Jyoti K, Baunthiyal M, Singh A (2016). Characterization of silver nanoparticles synthesized using Urtica dioica Linn. Leaves and their synergistic effects with antibiotics. Journal of Radiation Research and Applied Sciences 9(3):217-227.
Crossref
|
|
|
|
|
Kalimuthu K, Babu RS, Venkataraman D, Bilal M, Gurunathan S (2008). Biosynthesis of silver nanocrystals by Bacillus licheniformis. Colloids and Surfaces B: Biointerfaces 65(1):150-153.
Crossref
|
|
|
|
|
Kannan R, Berger C, Myneni S, Technau GM, Shashidhara LS (2010). Abdominal-A mediated repression of Cyclin E expression during cell-fate specification in the Drosophila central nervous system. Mechanisms of Development 127(1-2):137-145.
Crossref
|
|
|
|
|
Keshavamurthy M, Srinath BS, Rai VR (2017). Phytochemicals mediated green synthesis of gold nanoparticles using Pterocarpussantalinus L. (Red Sanders) bark extract and their antimicrobial properties. Particulate Science and Technology 36(7):785-790.
Crossref
|
|
|
|
|
Kumar CG, Mamidyala SK (2012). Extracellular synthesis of silver nanoparticles using culture supernatant of Pseudomonas aeruginosa. Colloids and Surfaces B: Biointerfaces 84(2):462-466.
Crossref
|
|
|
|
|
Manivasagan P, Venkatesan J, Senthilkumar K, Sivakumar K, Se-Kwon, K (2013). Biosynthesis, Antimicrobial and Cytotoxic Effect of SilverNanoparticles Using a Novel Nocardiopsis sp. MBRC-1. BioMed Research International 1:1-9.
Crossref
|
|
|
|
|
Matsumura Y, Yoshikata K, Kunisaki S, Tsuchido T (2003). Mode of bactericidal action of silver zeolite and its comparison with that of silver nitrate. Applied and Environmental Microbiology 69(7):4278-4281.
Crossref
|
|
|
|
|
Mohammadi F, Yousefi M, Ghahremanzadeh R (2019). Green Synthesis, Characterization and Antimicrobial Activity of Silver Nanoparticles (AgNPs) Using Leaves and Stems Extract of Some Plants. Advanced Journal of Chemistry 2(4):266-275.
Crossref
|
|
|
|
|
Mohanpuria P, Rana KN, Yadav SK (2008). Biosynthesis of nanoparticles: Technological concepts and future applications. Journal of Nanoparticle Research 10:507-517.
Crossref
|
|
|
|
|
Morones JR, Elechiguerra JL, Camacho A, Holt K, Kouri JB, Ramfrez, JT, Yacaman M (2005). The bactericidal effect of silver nanoparticles. Journal of Nanotechnology 16(10):2346-2353.
Crossref
|
|
|
|
|
Narayanan KB, Sakthivel N (2010). Biological synthesis of metal nanoparticles by microbes. Advances in Colloid and Interface Science 156(1-2):1-13.
Crossref
|
|
|
|
|
Ozcelika B, Orhanb I, Tokerb G (2006). Antiviral and antimicrobial assessment of some selected flavonoids. Biological Activity of Flavonoids 61(9-10):632-638.
Crossref
|
|
|
|
|
Pal S, Tak YK, Song JM (2007). Does the Antibacterial Activity of Silver Nanoparticles Depend on the Shape of the Nanoparticle? A Study of the Gram-Negative Bacterium Escherichia coli. Applied and Environmental Microbiology 27(6):1712-1720.
Crossref
|
|
|
|
|
Prabhu S, Poulose E (2012). Silver nanoparticles: mechanism of antimicrobial action, synthesis, medical applications, and toxicity effects. International Nano Letters 2(1):1-10.
Crossref
|
|
|
|
|
Saha S, Sarkar J, Chattopadhayay D, Patra S, Chakraborty A, Acharya K (2010). Production of silver nanoparticles by a phytopathogenic fungus Bipolaris nodulosa and its antimicrobial activity. Digest Journal of Nanomaterials and Biostructures 5(4):887-895.
|
|
|
|
|
Saklani V, Suman JV, Jain K (2012). Microbial synthesis of silver nanoparticles: A Review. Journal of Biotechnology and Biomaterials S13:007.
Crossref
|
|
|
|
|
ŠedÄ›nková I, Trchová M, Stejskal J, Prokeš J (2009). Solid-state reduction of silver nitrate with polyaniline base leading to conducting materials. ACS Applied Materials and Interfaces 1(9):1906-1912.
Crossref
|
|
|
|
|
Sharma N, Pinnaka AK, Raje M, Ashish FN, Bhattacharyya MS, Choudhury AR (2012). Exploitation of marine bacteria for production of gold nanoparticles. Microbial Cell Factories 11:86.
Crossref
|
|
|
|
|
Singh AK, Tiwari R, Kumar V, Singh P, Khadim SKR, Tiwari A, Srivastava V, Hasan SH, Asthana RK (2017). Photo-induced biosynthesis of silver nanoparticles from aqueous extract of Dunaliella salina and their anticancer potential. Journal of Photochemistry and Photobiology B: Biology 166:202-211.
Crossref
|
|
|
|
|
Singh C, Sharma V, Naik PK, Khandelwal V, Singh H (2011). Green biogenic approach for synthesis of gold and silver nanoparticles using Zingiber officinale. Digest Journal of Nanomaterials and Biostructures 6(2):535-542.
|
|
|
|
|
Sondi I, Salopek-Sondi B (2004). Silver nanoparticles as antimicrobial agent: a case study on E. coli as a model for Gram-negative bacteria. Journal of Colloid and Interface Science 275(1):17-182.
Crossref
|
|
|
|
|
Sunkar S, Nachiyar CV (2012). Microbial Synthesis and Characterization of Silver Nanoparticles Using the Endophytic Bacterium Bacillus cereus: A Novel Source in the Benign Synthesis. Global Journal of Medical Research 12(2):1-9.
|
|
|
|
|
Thu TT, Thi THV, Thi HN (2013). Biosynthesis of silver nanoparticles using Tithonia diversifolia leaf extracts and their antimicrobial activity. Materials Letters 105:220-223.
Crossref
|
|
|
|
|
Vanmathi SK, Sivakumar T (2012). Isolation and characterization of silver nanoparticles from Fusarium oxysporum. International Journal of Current Microbiology and Applied Sciences 1(1):56-62.
|
|
|
|
|
Venkatesan KR, Vajrai R, Nithyadevi M, Arun KP, Uma MK, Brindham P (2013). Characterization and Antimicrobial effect of silver Nanoparticles Synthesized from Bacillus subtilis (MTCC 441). International Journal of Drug Development and Research 5(4):187-193.
|
|
|
|
|
Vishwanatha T, Keshavamurthy M, Mallappa M, Murugendrappa MV, Nadaf YF, Siddalingeshwara KG, Dhulappa A (2018). Biosynthesis, characterization, and antibacterial activity of silver nanoparticles from Aspergillus awamori. Journal of Biology and Biotechnology 6(5):12-16.
Crossref
|
|
|
|
|
Walker LR (2003). Colonization dynamics and facilitative impacts of a nitrogen-fixing shrub in primary succession. Journal of Vegetation Science 14(2):277-290.
Crossref
|
|
|
|
|
Wolfe BE, Klironomos JN (2005). Breaking new ground: Soil communities and exotic plant invasion. BioScience 55(6):477-487.
Crossref
|
|
|
|
|
Yamal G, Sharmila P, Rao KS, Pardha S (2013). In built potential of YEM medium and its constituents to generate Ag/Ag2O nanoparticles. PLoS One 8:e61750.
Crossref
|
|