ABSTRACT
This study was conducted to determine the in-vitro probiotic properties of Enterococcus faecium strains isolated from soft cheese. To evaluate the safety of Enterococcus strains, we compared the pathogenic genes, antimicrobial susceptibility of the probiotic strains to those of clinical isolates, and their antimicrobial activity against food-borne pathogenic and spoilage bacteria. Enterococcus strains were identified and evaluated in vitro for biochemistry methods acid, bile salts, lysozyme and pancreatin tolerance. One hundred and three strains were identified as E. faecium, and none of them were no vancomycin-resistant, and no pathogenic genes – such as cylA, asa1, gelE, ace and cpd – were found. The isolates showed good viability at 120 and 240 min of incubation with pH 3.0, and were able to resist 0.3% and 0.1 g/ml of bile salts and pancreatic enzyme, respectively. One observed strong autoaggregation phenotype, and the isolates demonstrated high activity against L. innocua, L. monocytogenes, E. faecalis S. aureus, Salmonella Enteritidis and Salmonella Typhimurium. The results instigate the continuity of studies of E. faecium isolates in order to obtain a known probiotic strain.
Key words: Enterococcus, good bacteria, pathogenic genes, foods, antimicrobial activity.
The use of Enterococcus spp. in the making of fermented foods, such as milk, yogurt, cheese, fermented sausages and vegetables (Foulquié Moreno et al., 2006) has a long record in the history of food. Selected Enterococcus strains have been employed as probiotics in the promotion of both human and animal health ,improving the intestinal microbial balance (Foulquié Moreno et al., 2006; Franz et al., 2011) and producing enterocins (antimicrobial peptides) to inhibit the growth of food-borne pathogenic and spoilage bacteria (Ogaki et al., 2016).
Other therapeutic or prophylactic properties associated with probiotic enterococci include the improvement of constipation and diarrhea, reduction in cholesterol levels, stimulation of immunity and suppression of the carcinogenesis (Agerholm-Larsen et al., 2000; de Roos and Katan, 2000; Parvez et al., 2006; Meurman and Stamatova, 2007; Candela et al., 2008).
However, presence of enterococci in foods may present conflicting effects, either as a risk, a foreign (?) or as an indicator of poor hygiene during the processing of food (Bhardwaj et al., 2008). Some types of Enterococcus produce virulence factors (Jett et al., 1994; Foulquié Moreno et al., 2006), and are sometimes associated with pathogenicity (Khan et al., 2010). They have been reported to be the cause of endocarditis, bacteraemia, and several infections, besides multiple antibiotic resistances (Kayser, 2003). In addition, vancomycin-resistant Enterococcus (VRE) emerged and has become a major public health problem in several countries (Foulquié Moreno et al., 2006).
One cannot presume whether a specific probiotic bacterium will have a beneficial effect on health, except through determination of its genus or species. Reports on the safety of probiotics are limited, and there are few details about the nature of probiotic bacterial species (Sanders et al., 2010; Fijan, 2014). As part of the selection of new probiotic enterococci candidates, one needs to do a series of in vitro and in vivo analyses to assess their probiotic properties. Carrying no virulence factors nor vancomycin-resistant genes is a prior condition to regard an enterococci candidate as safe and eligible to be used as a starter of cultures, co-cultures; on the other hand, the probiotics that are acceptable for the preparation of food and medicines for humans are those which occur naturally in the intestinal tract of healthy human subjects and foods (Sanders et al., 2010).
Other criteria for potential probiotic strains should include their ability to colonize the intestinal tracts of humans and other mammals (Verschuere et al., 2000), and their resistance to survive humans’ biological barriers, such as the strains that have proven ability to survive the gastrointestinal tract (Dunne et al., 2001; Vinderola and Reinheimer, 2003), the presence of proteolytic enzymes and low pH values, bile salts and pancreatic juices.
Probiotic cultures should also be antagonistic to pathogenic bacteria by producing antimicrobial substances and must be safe for human use, maintaining their viability and beneficial properties during manu-facturing processes (Schillinger et al., 2005).
Therefore, the objective of this investigation was to perform a characterization of new food enterococcal strains of cheese origin and elicit their potential application as probiotics.
Bacterial strains and culture preparation
The study comprised one hundred and three Enterococcus spp. strains isolated from artisanal soft cheeses. Such isolates were identified as members of the Enterococcus spp. based on the phenotypic and genotypic criteria as previously reported (Furlaneto-Maia et al., 2014). A single probiotic culture containing strain Lactobacillus acidophilus LA-5 was used as control (Chr. Hansen). The bacterial strains were reactivated in MRS (Merck, Darmstadt, Germany) broth medium for 18 h at 37ºC under shaking conditions. Cells were harvested by centrifugation at 10000 g for 5 min and washed twice in NaCl solution (0.85% w/v). The pellet was resuspended in physiologic solution in order to obtain a suspension that contained approximately 109-1010 CFU/mL.
Antimicrobial susceptibility testing
Antibiotic discs (Laborclin®) were used to determine the strains susceptibility to ampicillin (AMP, 10 µg), nalidixic acid (NAL, 30 µg), vancomycin (VAN, 30 µg), erythromycin (ERY, 15 µg), chloramphenicol (CLO, 30 µg), norfloxacin (NOR, 10 µg), tetracycline (TET, 30 µg), imipenem (IPM 10 µg), amikacin (AK, 30 µg); cephalothin (CF, 30 µg); ciprofloxacin (CIP 5 µg); amoxicillin/clavulanic acid (AMC, 30 µg). The discs were placed onto Mueller–Hinton agar plates overlayed with the enterococcal culture with cell concentration corresponding to 0.5 McFarland standard turbidity. After incubation at 37°C for 18-24 h, the diameter of inhibition haloes around the colonies was measured. Susceptibility or resistance was interpreted in accordance with the Clinical Laboratory Standard Institute (CLSI, 2011) recommendations, and Staphylococcus aureus 25923 ATCC were used as strain quality control.
Determination of virulence factors
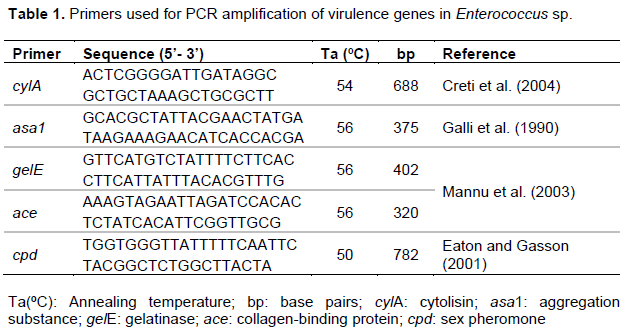
Enterococcus spp. genomic DNA was extracted by boiling method (Furlaneto-Maia et al., 2014). Determination of virulence factors was performed using a polymerase chain reaction (PCR) method. PCR assay was carried out using species-specific primers (Table 1). All PCR amplifications were performed in a final volume of 20 μl containing 1 ρmol of each primer (Forward e Reverse), 0.17 mM dNTPs, 2.5 mM MgCl2, 1 U of Taq DNA polymerase (Invitrogen), buffer of Taq, and 10 µl template DNA. One observed an initial cycle of denaturation (94°C for 2 min), followed by 30 cycles of denaturation (94°C for 1 min), annealing at an appropriate temperature (Table 1) for 1 min and elongation (72°C for 10 min). A thermal cycler (Techne-Tc3000) was used to perform the PCR reactions. PCR products were analysed by gel electrophoresis in 1.5% agarose stained with ethidium bromide (0.5 g.ml-1), observed under UV transillumination and photographed with L-PIX ST (LOCCUS).
Hemolytic activity
To investigate the production of hemolysin, the strains grown in MRS broth were streaked onto layered agar plates with 7% v/v fresh sheep blood (Himedia), then grown at 37°C for 48 h. β-hemolysis was revealed by the formation of clear zones surrounding the colonies on the blood agar plates (Foulquié Moreno et al., 2006).
Effects of low pH on growth rate
The effects of low pH on growth rate were determined as previously described by Oluwajoba et al. (2013), with modifications. Enterococcus spp. bacterial colonies were incubated for 0, 1, 2, 3 and 4 h at 37ºC in MRS medium, then adjusted to pH 3 with HCl (4 mol/l). The number of CFU/ml was calculated and compared to the CFU/ml at time 0. The surviving bacteria were counted on the MRS agar, and all these experiments were performed in triplicate.
Lysozyme, bile salts and pancreatin resistance
To simulate the saliva in vitro, 200 µL of the bacterial suspensions were inoculated in a sterile electrolyte solution-SES (0.22 g/L CaCl2, 6.2 g/L NaCl, 2.2 g/L KCl, 1.2 g/L NaHCO3) in the presence of 100 mg/L of lysozyme (Sigma-Aldrich) in accordance with Vizoso-Pinto et al. (2006). Bacterial suspensions in SES without lysozyme were included as control. Samples were incubated at 37ºC and microbial counts after 0, 30 and 120 min were carried out on MRS agar (24-48 h; 37ºC). Survival rate was calculated as percentage of the CFU/mL after 30 and 120 min in comparison to the CFU/mL at time 0.
Resistance to bile salts and pancreatin was measured as described by Charteris et al. (1998), with modifications. The overnight culture was adjusted to pH 8 and a solution of bile salts (Oxoid) was added to a final concentration of 0.3% or 0.1 g/ml of Pancreatin (Sigma). The mixture (bile salt/ bacterial cells and pancreatin/bacterial cells) was incubated for 0 and 240 min at 37°C. Aliquots were taken for determination of CFU onto the MRS agar. The plates were incubated for 48 h. The addition of bile salt was omitted in the control tube. Results were expressed as percentage of growth as compared to the control (CFU/mL at time zero).
Autoaggregation and co-aggregation assay
The extent of autoaggregation and co-aggregation in the selected probiotic isolates was assessed with the method described by Kos et al. (2003), and the percentage of autoaggregation and co-aggregation was calculated by following Mojgani et al. (2015) descriptions. As to the autoaggregation, overnight-grown cultures of the tested isolates were harvested by centrifugation and the pellet was suspended in PBS (pH 7.0) to obtain an OD (600 nm) of 0.6. The tubes were incubated at 37°C, and the absorbance at 600 nm of the celular suspensions was monitored every 1 h for a period of 5 h. Co-aggregation assay was performed by mixing equal volumes of a washed-cell suspension of selected probiotic isolates with equal volume of overnight grown cultures of L. monocytogenes (CDC 4555). The tubes were incubated at room temperature and absorbance at 600 nm was measured at 5 h. Controls included pure cultures of bacterial cells suspension in PBS.
Screening for enterocin production
The antimicrobial screening assay was evaluated in accordance with Ogaki et al. (2016). Enterococci strains were streaked in plates containing MRS agar, which were then incubated for 24 h at 37°C. The plates were inverted to receive 1 mL of chloroform in the covers, and remained closed for 20 min. Residual chloroform was evaporated by opening the plates. Using the pour plate method, each indicator strain (108 cells.mL-1) was inoculated into soft MRS agar (0.8%), poured into plates forming an overlay, and these plates were incubated for 24 h at 37°C. If inhibitions zones were found around the colonies, the isolates were considered able to produce enterocin. One used indicator strains such as Listeria innocua CLIP 12612, L. monocytogenes CDC 4555, Enterococcus faecalis ATCC 29212, S. aureus ATCC 25925, S. aureus ATCC 29213, S. aureus ATCC 6538, Salmonella Enteritidis ATCC 13076, Salmonella Typhimurium UK1 and Escherichia coli BAC 49LT ETEC.
Statistical analysis
Statistical analysis was carried out using the software STATISTICA 7 (StatSoft Italia, Padova, Italy). Analysis of variance test (ANOVA) was done in order to determine a significant difference of viability among Enterococcus strains and L. acidophilus. The collected data were analysed at the significance level of p < 0.05.
Of all strains, 53 were chosen based on their absence of virulence, hemolysis and antimicrobial susceptibility. Almost 2% of the strains showed resistance to vancomicyn and eritromicyn, and 54% to tetracyclin, while other strains were sensitive to all antimicrobial used.
Twenty-four strains (that is, 45%) were in-vitro resistant to bile salt and pancreatic enzyme, ranging from a minimum value of 81.5% to a maximum of 105 and 79.2% to 108.2, respectively (Table 2). The low pH-tolerance property of 24 Efm strains was investigated by culturing at pH 3.0 for 120 and 240 min. Of these, seven strains showed higher tolerance, with a survival rate greater than the control strain (L. acidophilus) (Table 2), in particular, the Efm 55, Efm 58, Efm 67, Efm 9A, Efm 16A, Efm 19A, Efm 44A strains.
In addition, it was studied the survival of these isolates in SES solution containing 100 mg/ml of lysozyme. The isolated strains survived in the presence of lysozyme for 30 and 120 min.
When taken together, results showed that strains Efm 55, Efm 58, Efm 67, Efm 9A, Efm 16A, Efm 19A, Efm 44A were significantly different (p < 0.05) in all conditions as compared with the control strain.
According to the autoaggregation results, the Efm9A, Efm19A and Efm67 strains demonstrated strong auto-aggregation phenotype, 100, 92 and 50%, respectively, within 5 h of incubation. Moreover, the Efm55, Efm58 strains showed moderate autoaggregation values (45-37%), and the Efm16A did not show any aggregation during the incubation hour. All strains exhibited co-aggregation with strain-pathogen (L. monocytogenes), showing values among 65 to 78%.
The antimicrobial spectra of Enterococcus strains were investigated by using 9 pathogens as targets. The isolated strains demonstrated broad activity against all tested Gram-positive (L. innocua CLIP 12612, L. monocytogenes CDC 4555, E. faecalis ATCC 29212, S. aureus ATCC 25925, S. aureus ATCC 29213, S. aureus ATCC 6538) and Gram-negative (S. Enteritidis ATCC 13076, S. Typhimurium UK1) strains, with halos ranging from 0.4 to 1.52 mm. Although that E. coli BAC 49LT ETEC was not inhibited by Enterococcus strains.
Among the Lactic Acid Bacteria (LAB), members of the Enterococcus genus have been object of increasing scientific work, because of its wide range of health-promoting effects. The commonly accepted criteria is that probiotic organisms should be resistant to acid and bile, which are elements present in the stomach and small intestine conditions. In our previous work, the E. faecium demonstrated high ability to survive in the presence of lysozyme and pancreatic enzymes, bile salt and low pH, during several hours. More importantly, none of the E. faecium strains carried the virulence factors cylA and cylB, required in hemolytic activity, which is the most important virulence trait that lyses the eukaryotic cells (Kayser, 2003). E. faecium also showed low antimicrobial resistance, though antimicrobial-resistant probiotics can be used in combination with antimicrobial agents (Sanders et al., 2010).
Based on cell growth /survival, we selected seven E. faecium strains for investigation. These strains, initially named as Efm 55, Efm 58, Efm 67, Efm 9A, Efm 16A, Efm 19A, Efm 44A, presented significant activity when compared with the control bacteria.
E. faecium is found in many food products, especially those from animal origin, such as dairy products (Foulquié Moreno et al., 2006; Kivanç et al., 2016). They are most frequently present in many traditional cheeses – prepared mostly from raw ewes’ or goats’ milk –, and play an important role in the ripening of such products (Manolopoulou et al., 2003). A high prevalence of enterococci in processed foods may be attributed to their resistance to heat, extreme salinity and harsh conditions during the ripening of fermented foods (Gomes et al., 2008; Jurkovic et al., 2006). Altogether, enterococci strains have been a promising probiotic in the promotion of human and animal health by improving the intestinal microbial balance (Foulquié-Moreno et al., 2006; Franz et al., 2011; Buntin et al., 2008).
In this study, Efm strains were exposed to pH 3.0 for 240 min, and several strains were highly resistant to pH 3.0 with levels that were higher than the control bacteria. The average time food stays in the stomach is 3 h, and, in general, our results meet those of other researchers (Mansour et al., 2014).
Once bacteria have survived the gastric barrier (low pH), the environment in the small intestine is a second major barrier for probiotic strains. Therefore, authors have recommended testing bacterial resistance to bile salt concentrations in the 0.3% and pancreatin 0.1 mg/mL to the selection of probiotic bacteria for human use (Bezkorovainy, 2001; Tuomola et al., 2001; Mansour et al., 2014). The major factors determining the survival of LAB include particular characteristics of the strains, tolerance to acid and bile, and resistance to gastric and intestinal juices (Succi et al., 2005). Amaral et al. (2017) and Sun et al. (2010) showed that E. faecium was more stable during the simulation of the gastrointestinal tract, showing greater cell viability.
High acidity and high concentration of bile components in the gastrointestinal tract influence the selection of potential probiotic strains (Hyronimus et al., 2000). However, small intestine tolerance is potentially more important than gastric survival. With the development of new delivery systems and the use of specific foods, some studies indicate that acid-sensitive strains can be buffered through the stomach. However, in order to promote a positive effect in the host, probiotics need to survive and colonize his/her small intestine, and the condition of such environment may be an essential criterion for future probiotics (Huang and Adams, 2004).
This study investigated the antibacterial activity of E. faecium strains isolated from soft cheese. These E. faecium strains were able to inhibit L. innocua, L. monocytogenes, E. faecalis, S. aureus, and Salmonella. In particular, E. coli was not sensitive to all E. faecium strains.
Besides determining that enterococci strains showed high auto-aggregation, one has also demonstrated that they exhibit high co-aggregation against L. monocytogenes strain. Aggregation and co-aggregation among bacteria play an important role in the prevention of surface-colonization by pathogens (García-Cayuela et al., 2014), as it is well known that the co-aggregation abilities of LAB strains might interfere with the ability of pathogenic species to infect the host, and can also prevent the colonization of food-borne pathogens (García-Cayuela et al., 2014).
In summary, the results obtained in this study suggest that E. faecium strains are resistant to pass through the gastrointestinal tract. One also verified the viability of this strain through the exposure rate and the combination of simulated gastric juice and bile salts, intestinal juice, bile and acid tolerance. Further investigations may be warranted to elucidate its potential health benefit and its application as a promising probiotic strain in the food industry.
This study have demonstrated that E. faecium strains of soft-cheese origin may be a probiotic candidate with functional characteristics in terms of resistance to low pH and bile salts, survival under digestion conditions and adhesion, antimicrobial properties, antibiotic resistance, and presence of the virulence factors as well as hemolytic reaction. Further work is in progress to characterize both the bacteriocin(s) and its probiotic functionality.
The authors have not declared any conflict of interests.
REFERENCES
|
Agerholm-Larsen L, Bell ML, Grunwald GK, Astrup A (2000). The effect of a probiotic milk product on plasma cholesterol: a meta-analysis of short-term intervention studies. Eur. J. Clin. Nutr. 54(11):856-860.
Crossref
|
|
|
|
Amaral DM, Silva LF, Casarotti SN, Nascimento LC, Penna AL (2017). Enterococcus faecium and Enterococcus durans isolated from cheese: Survival in the presence of medications under simulated gastrointestinal conditions and adhesion properties. J. Dairy Sci. 100(2):933-949.
Crossref
|
|
|
|
|
Bezkorovainy A (2001). Probiotics: determinants of survival and growth in the gut. Am J. Clin. Nutr. 73:399-405.
|
|
|
|
|
Bhardwaj A, Malik RK, Chauhan P (2008). Functional and safety aspects of enterococci in dairy foods. Indian J. Microbiol. 48(3):317-
Crossref
|
|
|
|
|
Buntin N, Chanthachum S, Hongpattarakere T (2008). Screening of lactic acid bacteria from gastrointestinal tracts of marine fish for their potential use as probiotics. Songklanakarin J. Sci. Technol. 30:141-148.
|
|
|
|
|
Candela M, Miccoli G, Bergmann S, Turroni S, Vitali B, Hammerschmidt S, Brigidi P (2008). Plasminogen-dependent proteolytic activity in Bifidobacterium lactis. Microbiol. 154:2457–2462.
Crossref
|
|
|
|
|
Charteris WP, Kelly PM, Morelli L, Collins JK (1998). Development and application of an in vitro methodology to determine the transit tolerance of potentially probiotic Lactobacillus and Bifidobacterium species in the upper human gastrointestinal tract. J. Appl. Microbiol. 84:759-768.
Crossref
|
|
|
|
|
CLSI (Clinical and Laboratory Standards Institute) (2011). Performance Standards for Antimicrobial Susceptibility Testing; 20th Informational Supplement Institute Wayne, PA: M100-S21:31.
|
|
|
|
|
Creti R, Imperi M, Bertuccini L, Fabretti F, Orefici G, Di Rosa R, Baldassarri L (2004). Survey for virulence determinants among Enterococcus faecalis isolated from different sources. J. Med. Microbiol. 53:13-20.
Crossref
|
|
|
|
|
de Roos NM, Katan MB (2000). Effects of probiotic bacteria on diarrhea, lipid metabolism, and carcinogenesis: a review of papers published between 1988 and 1998. Am. J. Clin. Nutr. 71(2):405-411.
|
|
|
|
|
Dunne C, O'Mahony L, Murphy L, Thornton G, Morrissey D, O'Halloran S, Feeney M, Flynn S, Fitzgerald G, Daly C, Kiely B, O'Sullivan GC, Shanahan F, Collins JK (2001). In vitro selection criteria for probiotic bacteria of human origin: correlation with in vivo findings. Am. J. Clin. Nutr. 73(2):386s-392s.
|
|
|
|
|
Eaton TJ, Gasson MJ (2001). Molecular screening of Enterococcus virulence determinants and potential for genetic exchange between food and medical isolates. Appl. Environ. Microbiol. 67(4):1628-1635.
Crossref
|
|
|
|
|
Fijan S (2014). Microorganisms with claimed probiotic properties: An overview of recent literature. Int. J. Environ. Res. Public Health. 11:4745-4767.
Crossref
|
|
|
|
|
Foulquié Moreno MR, Sarantinopoulos P, Tsakalidou E, De Vuyst L (2006). The role and application of enterococci in food and health. Int. J. Food Microbiol. 106(1):1-24.
Crossref
|
|
|
|
|
Franz CMAP, Huch M, Abriouel H, Holzapfel W, Gálvez A (2011). Enterococci as probiotics and their implications in food safety. Int. J. Food Microbiol. 151(2):125-140.
Crossref
|
|
|
|
|
Furlaneto-Maia L, Rocha KR, Henrique FC, Giazzi A, Furlaneto MC (2014). Antimicrobial Resistance in Enterococcus sp Isolated from Soft Cheese in Southern Brazil. Adv. Microbiol. 4(3):175-181.
Crossref
|
|
|
|
|
Galli D, Lottspeich F, Wirth R (1990). Sequence analysis of Enterococcus faecalis aggregation substance encoded by the sex pheromone plasmid pAD1. Mol. Microbiol. 4(6):895–904.
Crossref
|
|
|
|
|
García-Cayuela T, Korany A M, Bustos I, de Cadi-anos LPG, Requena T, Peláez C, Martínez-Cuesta, MC (2014). Adhesion abilities of dairy Lactobacillus plantarum strains showing an aggregation phenotype. Food Res. Int. 57:44-50.
Crossref
|
|
|
|
|
Gomes BC, Esteves CT, Palazzo IC, Darini AL, Felis GE, Sechi LA, Franco BD, De Martinis EC (2008). Prevalence and characterization of Enterococcus spp. isolated from Brazilian foods. Food Microbiol. 25(5):668-675.
Crossref
|
|
|
|
|
Huang Y, Adams MC (2004). In vitro assessment of the upper gastrointestinal tolerance of potential probiotic dairy propionibacteria. Int. J. Food Microbiol. 91(3):253-260.
Crossref
|
|
|
|
|
Hyronimus B, Le Marrec C, Hadj Sassi A, Deschamps A (2000). Acid and bile tolerance of spore-forming lactic acid bacteria. Int. J. Food Microbiol. 61:193-197.
Crossref
|
|
|
|
|
Jett B D, Huycke MM, Gilmore MS (1994). Virulence of enterococci. Clin. Microbiol. Rev. 7(4):462-478.
Crossref
|
|
|
|
|
Jurkovic D, Krizková L, Dusinskyý R, Belicová A, Sojka M, Krajcovic J, Ebringer L (2006). Identification and characterization of enterococci from bryndza cheese. Lett. Appl. Microbiol. 42(6):553-559.
Crossref
|
|
|
|
|
Kayser FH (2003). Safety aspects of enterococci from the medical point of view. Int. J. Food Microbiol. 88:255-262.
Crossref
|
|
|
|
|
Khan H, Flint S, Yu PL (2010). Enterocins in food preservation. Int. J. Food Microbiol. 141:1-10.
Crossref
|
|
|
|
|
Kivanç SA, Kivanç M, Yigit T (2016). Antibiotic susceptibility, antibacterial activity and characterisation of Enterococcus faecium strains isolated from breast milk. Exp Ther Med. 12(3):1732–1740.
|
|
|
|
|
Kos B, Susković J, Vuković S, Simpraga M, Frece J, Matosić S (2003). Adhesion and aggregation ability of probiotic strain Lactobacillus acidophilus M92. J. Appl. Microbiol. 94(6):981-987.
Crossref
|
|
|
|
|
Mannu L, Paba A, Daga, E, Comunian R, Zanetti S, Duprè I, Sechi LA (2003). Comparison of the incidence of virulence determinants and antibiotic resistance between Enterococcus faecium strains of dairy, animal and clinical origin. Int. J. Food Microbiol. 88:291-304.
Crossref
|
|
|
|
|
Manolopoulou E, Sarantinopoulos P, Zoidou E, Aktypis A, Moschopoulou E, Kandarakis IG, Anifantakis EM (2003). Evolution of microbial populations during traditional Feta cheese manufacture and ripening. Int. J. Food Microbiol. 82(2)153-161.
Crossref
|
|
|
|
|
Mansour NM, Heine H, Abdou SM, Shenana ME, Zakaria MK, El-Diwany A (2014). Isolation of Enterococcus faecium NM113, Enterocccus faecium NM213 and Lactobacillus casei NM512 as novel probiotics with immunomodulatory properties. Microbiol. Immunol. 58(10):559-569.
Crossref
|
|
|
|
|
Meurman JH, Stamatova I (2007). Probiotics: Contributions to oral health. Oral Dis. 13(5):443-451.
Crossref
|
|
|
|
|
Mojgani N, Hussaini F, Vaseji N (2015). Characterization of indigenous Lactobacillus strains for probiotic properties. Jundishapur J. Microbiol. 8(2):e17523.
Crossref
|
|
|
|
|
Ogaki MB, Rocha KR, Terra MR, Furlaneto MC, Furlaneto-Maia L (2016). Screening of the Enterocin-encoding genes and antimicrobial activity in Enterococcus species. J. Microbiol. Biotechnol. 26(6):1026–1034.
Crossref
|
|
|
|
|
Oluwajoba SO, Akinyosoye FA, Oyetayo VO (2013). In vitro screening and selection of probiotic lactic acid bacteria isolated from spontaneously fermenting Kunu-Zaki Adv in Microbiol. 3:309-316.
Crossref
|
|
|
|
|
Parvez S, Malik KA, Ah Kang S, Kim HY (2006). Probiotics and their fermented food products are beneficial for health. J. Appl. Microbiol. 100(6):1171-1185.
Crossref
|
|
|
|
|
Sanders ME, Akkermans LMA, Haller D, Hammerman K, Heimbach J, Hörmannsperger G, Huys G, Levy DD, Lutgendorff F, Mack D, Phothirath P, Solano-Aguilar G, Vaughan E (2010). Safety assessment of probiotics for human use. Gut Microbes. 1(3):164-185.
Crossref
|
|
|
|
|
Schillinger U, Guigas C, Holzapfel WH (2005). In vitro adherence and other properties of lactobacilli used in probiotic yogurt-like products. Int. Dairy J. 15:1289-1297.
Crossref
|
|
|
|
|
Succi M, Tremonte P, Reale A, Sorrentino E, Grazia L, Pacifico S, Coppola R (2005). Bile salt and acid tolerance of Lactobacillus rhamnosus strains isolated from Parmigiano Reggiano cheese. FEMS Microbiol. Lett. 244(1):129-137.
Crossref
|
|
|
|
|
Sun P, Wang J, Jiang Y (2010). Effects of Enterococcus faecium (SF68) on immune function in mice. Food Chem. 123:63-68.
Crossref
|
|
|
|
|
Tuomola E, Crittenden R, Playne M, Isolauri E, Salminen S (2001). Quality assurance criteria for probiotic bacteria. Am. J. Clin. Nutr.73:393-398.
|
|
|
|
|
Verschuere L, Rombaut G, Sorgeloos P, Verstraete W (2000). Probiotic bacteria as biological control agents in aquaculture. Microbiol. Mol. Biol. Rev. 64(4):655–671.
Crossref
|
|
|
|
|
Vinderola CG, Reinheimer JA (2003). Lactic acid starter and probiotic bacteria:a comparative "in vitro" study of probiotic characteristics and biological barrier resistance. Food Res. Int. 36:895-904.
Crossref
|
|
|
|
|
Vizoso-Pinto MG, Franz CMAP, Schillinger U, Holzapfel W (2006). Lactobacillus spp. com in vitro propriedades probióticas de fezes humanas e produtos fermentados tradicionais. Int. J. Food Microbiol. 109:205-214
|
|