ABSTRACT
Yersinia enterocolitica, an important food-borne enteric pathogen is associated with various clinical manifestations ranging from self-limited gastroenteritis to more invasive syndrome such as terminal ileitis and mesenteric lymphadenitis. The main aim of this study was to investigate the incidence of Y. enterocolitica in milk samples. For this purpose, one hundred (100) samples of raw cow’s milk were collected from the western Algeria region. Seventeen (17) isolates were obtained. All these isolates belong to Y. enterocolitica biotype 1A and were ystB positive. Heat resistance and antibiotic susceptibility of these isolates were also investigated. The heat resistance D-value (decimal reduction time) and heat sensitivity ZT values (increase in temperature leads to a ten-fold reduction of the D value) of Y. enterocolitica in BHI broth showed that D55, D60 and D65 were 1.34, 0.85 and 0.62 min, respectively. The obtained Z value was 29.98°C and antibiotic resistance profiles of 17 isolates were evaluated. All the isolates were susceptible to 13 of the 30 tested antibiotic, resistance was noted for eight different antibiotics, among are them Ampicillin and 3rd generation Cephalosporins. The presence of chromosomal ystB gene virulence and antibiotic susceptibility indicate that these isolates from raw milk are potentially able to cause human foodborne illnesses and highlights the role of milk as a transmission vehicle of potentially pathogenic Y. enterocolitica strains, with consequent risks for consumer’s health via the consumption of raw milk and derivatives.
Key words: Yersinia enterocolitica, biotype, virulence gene, heat resistance, antibiotic resistance, raw milk.
Yersinia enterocolitica, which was first described in 1934 as a small Gram-negative coccobacillus psychro-tolerant enterobacterium, isolated from several environmental sources, that is, foods and human clinical samples are a causative organism in several out-breaks of gastroenteritis, in which foods were implicated (Bottone, 1999; Soltan-Dallal et al., 2004; Lambertz and Danielsson-Tham, 2005). In recent years, Y. enterocolitica has been the third most common cause of food borne diseases after Campylobacter spp. and Salmonella spp. (EFSA, 2011).
Y. enterocolitica has six biotypes, biotypes 1B, 2, 3, 4, and 5 which are known to be pathogenic and those of biotype 1A are considered as nonpathogenic (Bottone, 1999; Soltan-Dallal et al., 2004). The biotype 1A strains are generally regarded as non-virulent. They lack pYV plasmid and major chromosomal virulence genes. Despite this, some biotype 1A strains produce disease symptoms indistinguishable from that produced by known pathogenic biotypes (1B, 2-5). Some biotype 1A strains are able to invade epithelial cells, resist macrophages and carry genes associated with virulence (Tennant et al., 2003; Bhagat and Virdi, 2010). The ystB gene is widely distributed in Y. enterocolitica biotype 1A strains where the production of Yersinia stable toxin Yst-b is the major contributor to diarrhea produced by biotype 1A strains (Singh and Virdi, 2004b).
The frequent association of Y. enterocolitica with raw milk (Bernardino-Varo et al., 2013) and the ability of this organism to grow in milk at refrigeration temperatures (Bari et al., 2011) have been well documented. Some Y. enterocolitica biotypes are considered as the major prevalent milk-borne pathogens (Bernardino-Varo et al., 2013); they are responsible for gastroenteritis and other syndromes in humans and animals (Huovinen et al., 2010; Singh and Virdi, 2004b). Thus, its control is important for the safety of refrigerated dairy products (Ye et al., 2014). Y. enterocolitica has been isolated from raw milk and pasteurized dairy products in several countries e.g. in the USA (Jayarao and Henning, 2001), China (Wang et al., 2010; Ye et al., 2014), Mexico (Bernardino-Varo et al., 2013), Brazil (Falcão et al., 2006), Iran (Soltan-Dallah et al., 2004; Rahimi et al., 2014; Jamali et al., 2015), India (Subha et al., 2009), Turkey (Guven et al., 2010), Nigeria (Okeke and Okwori, 2014), Egypt (Darwish et al., 2015) and other countries. In Algeria, the raw milk is still frequently consumed. National production of raw cow’s milk is estimated at 2.3 billion liters. Only a third of this quantity is integrated to the industrial plants (ITELV, 2012), therefore, the most important issue about Y. enterocolitica is its control in raw milk and derivatives.
A broad spectrum of antibiotics has been widely used in agriculture to treat infections and improve growth and feed efficiency in livestock and poultry (Mathew et al., 2007). The need to use antibiotics in the treatment of humans and animals may lead to the development of mechanisms resistance antibiotic, causing a growing risk to human and animal health (Perkowska et al., 2011). For this reason, the use of antibiotic growth promoters in animal production must be prohibited or controlled in each country (Singh and Virdi, 2004a). Moreover, Y. enterocolitica produces beta-lactamase (penicillinase and cephalosporinase) that make them naturally resistant strains to Penicillins and Cephalosporins first and second generations (Singh and Virdi, 2004a). Systematic monitoring of the susceptibility of bacterial strains, including Y. enterocolitica, must therefore be regarded as highly justified to ensure appropriate treatment of humans and to limit the spread of microorganisms’ drug resistance in animals (Perkowska et al., 2011).
Sampling
One hundred (100) raw cow’s milk samples were collected aseptically from cans and tanks at the level of the dock receipt of dairy plant “Giplait Mansourah” located in Tlemcen city (northwest of Algeria) Samples were taken in a 250 mL sterile container, then transported to the laboratory in ice boxes. The journey took 15 min. At the laboratory, samples were immediately processed.
Raw cow’s milk collected at the dairy is from different areas of Western Algeria: Ain-Temouchent, Ain-Youcef Amieur, Beni Mester, Mansourah, Ouled Mimoun, Remchi, Sabra, Sebdou, Sidi Bel-Abbès and Tlemcen. Milk was brought by farmers in the early morning in cans and refrigerated tank trucks. The study covers the period from January to October 2013. On average, 2 to 3 samples per week were used. The number of farmers is about 700 with an average of 8 cows/ breeder.
Isolation and isolates identification
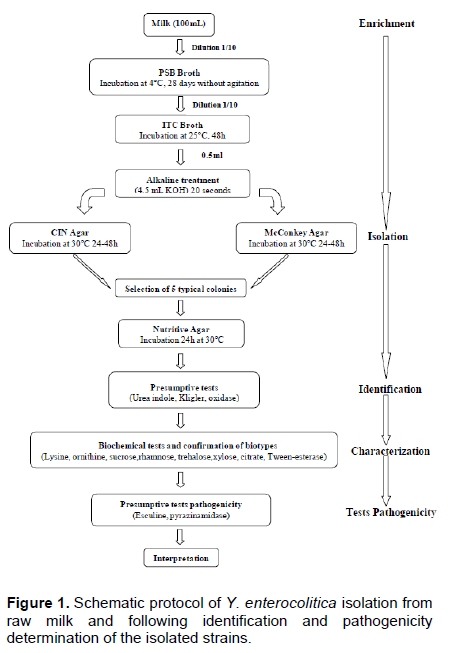
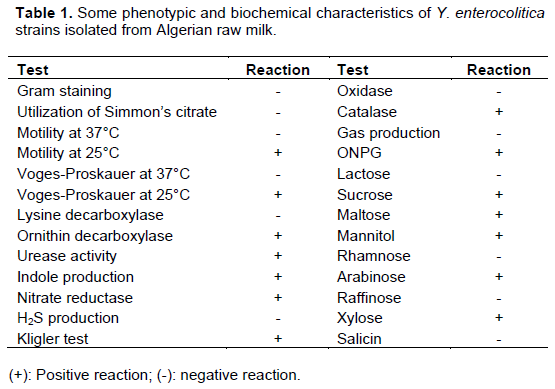
Y. enterocolitica strains were isolated using two enrichment steps, a pre-enrichment in Peptone Sorbitol Bile salts broth (PSB broth, Fluka, India) and an enrichment in Irgasan, Ticarcillin and potassium Chlorate broth (ITC broth, Fluka, India). One milliliter of each sample was added to 10 mL of PSB broth. The presumptive presence of Y. enterocolitica was checked after 4 weeks (28 days) of incubation at 4°C without shaking. One milliliter of each pre-enriched culture in PSB broth was added to 10 mL of ITC broth and incubated at 25°C for 48 h without shaking. In order to reduce the background contaminating flora, Aulisio’s alkali treatment method was performed: 0.5 mL of each enriched ITC broth was treated with 4.5 mL of 0.5% KOH solution (prepared in 0.5% NaCl solution), stirred for 20 s (AFNOR, 2003). Then, a loopful of the mixture was streaked immediately on Mac Conkey agar (Fluka, India) and Cefsulodin, Irgasan Novobiocin agar (CIN agar, Fluka, India) and incubated for 24 to 48 h at 30°C (Figure 1). The presumptive isolates were exanimate by biochemical tests as described by the ISO 10273:2003 horizontal method for the detection of presumptive pathogenic Y. enterocolitica with the following tests: Gram staining, oxidase, catalase, indole production, tryptophane deaminase, glucose and lactose fermentation, gas formation from glucose, H2S production, lysine decarboxylase, utilization of Simmons citrate, esculin hydrolysis, reduction of nitrate, mobility at 25 and 37°C and fermentation of xylose, mannitol and trehalose (Table 1) (AFNOR, 2003). The isolates were further identified by using the API 20E (BioMerieux, France). This system is still accepted as the good standard for the rapid identification of Y. enterocolitica (Tudor et al., 2008). The identification of biotype relies on a panel of biochemical tests as described in the ISO 10273-2003 method, allowing differen-tiation of pathogenic biotypes from the non-pathogenic biotype (AFNOR, 2003). The protocol of Y. enterocolitica isolation from raw milk and following identification and pathogenicity determination of the isolated strains is schematized in Figure 1.
Real-time PCR for detection of ystB gene
The PCR assays have been developed as an efficient tool for identifying pathogenic Y. enterocolitica (Lambertz and Danielsson-Tham, 2005). Real time PCR targeted the chromosomally-located ystB gene that is present in all Y. enterocolitica strains (Wang et al., 2010). Strains were sub-cultured on (plate count agar) PCA at 30°C for 24 h. DNA was extracted from colonies with QIAamp DNA mini kit (Qiagen, USA) following the manufacturer’s instructions. All PCR were performed on a CFX96 Real-Time PCR Detection System (Bio-Rad, Hercules, California), in a final volume of 25 μl with the Sybr® Green JumpstartTMTaqReadyMix TM (Sigma-Aldrich, Saint Louis, Missouri). The primers used in this study, were (5′-GTA CAT TAG GCC AAG AGA CG-3′) and (5′-GCA ACA TAC CTC ACA ACA CC-3′) (Baghat and Virdi, 2009). The final concentration of primers in the PCR reaction was 0.2 μM for ystB.
The PCR reaction was carried out under the following conditions: an initial denaturation at 95°C for 5 min, 34 cycles of 95°C for 10 s (denaturation), 55°C for 15 s (annealing) and 70°C for 1 min (extension) (Baghat and Virdi, 2009). The PCR products (146 bp) were visualized by ethidium bromide staining on 1.8% TBE agarose gel. Mass values are for 1 μg/lane. A 50- bp DNA ladder (Biolabs, New England) was used to determine the size of PCR product.
Heat treatment
One strain isolated from raw milk (baptized YHK261) was chosen for further characterization because of its unusual heat resistance. This strain was aseptically transferred to 100 mL Brain Heart Infusion Broth (BHI Conda, Spain) and incubated at 30°C for 24 h. 1 mL was sub-cultured in 100 mL BHI at 30°C for 18 h. At the stationary phase, cells were recovered and the culture was adjusted to a final colony count of 108 CFU mL-1. Heating temperatures of 55, 60 and 65°C were chosen based on previous studies reported in literature (Pagán et al., 1999). The vials containing 100 mL of sterile BHI were placed in water-bath heated at the preselected temperature (e.g. 55°C). The vials are fixed so that the broth is totally submerged in the bath. The sample temperature during treatment was monitored using a thermometer (IsoLab GmbH, German) placed in another vial containing 100 mL of BHI simultaneously placed in the water bath with the first one, and to minimize any risk of contamination. In a first step, a bacterial suspension was introduced into the heating medium. Samples were removed periodically and immediately placed in ice- water. Each cooled sample was serially diluted in 9 mL of sterile physiological saline (0.9% NaCl w/v). During the third step, dilutions cascade was performed. Direct counts were obtained by plating in duplicate from the dilution series onto trypticase soy agar (TSA Conda, Spain). After incubation at 30°C for 24 h, plates were examined for typical colonies of Y. enterocolitica. The number of colony forming units (CFU) on agar plates was converted to log10 CFU g-1. Each experiment was carried out in duplicate at each temperature.
The slope was obtained for each plot of log10 of surviving cells mL-1 against time using linear regression analysis Log10 D (T). The estimate of thermal resistance was obtained by fitting the linear regressions of the log10 number of surviving cells at each time interval. D values are the absolute value of the inverse slope of the regression line. These D values, in minutes, were used to fit plots of log10 D value versus temperature. To fit the models to the experimental data, the GraphPad PRISM (GraphPad Software, San Diego, CA, USA) was used. D values for Y. enterocolitica were calculated using the average slope for a given treatment. The value of the inverse slope obtained by plotting log10 D value versus temperature represents the Z value.
Antibiotic susceptibility testing
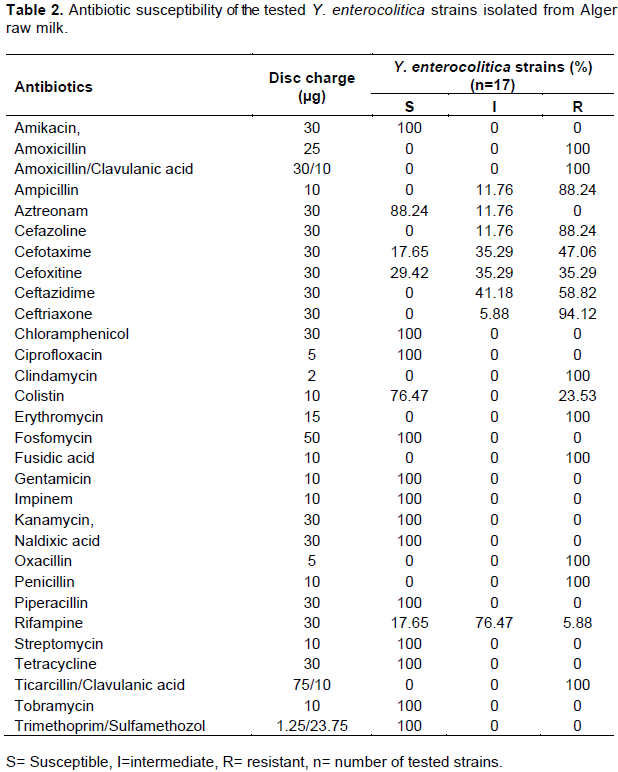
Y. enterocolitica isolates were examined for their susceptibility to b-lactam and non-b-lactam antibiotics. Antimicrobial susceptibility was determined by the standard disk diffusion method of Bauer, using Mueller-Hinton agar (Singh and Virdi, 2004a) and antibiotic disks were purchased from Pasteur Institute, Algeria. The plates were incubated (24 h at 37°C) and resistance was recorded via visual examination. Different antibiotics were tested (Table 2), including ampicillin (AM), amoxicillin (AMX), amoxicillin/clavulanic acid (AMC), oxacillin (OX), penicillin (P), ticarcillin/clavulanic acid (TCC), cefazolin (CZ), cefoxitin (FOX), ceftriaxone (CRO), ceftazidime (CAZ), cefotaxime (CTX), amikacin (AN), kanamycin (K), gentamicin (GM), tobramycin (TM), nalidixic acid (NA), ciprofloxacin (CIP), imipenem (IMP), trimethoprim/sulfamethozole (SXT), clindamycin (DA), colistin (CT), fusidic acid (FA), rifampin (RA), piperacillin (PI), aztreonam (ATM), chloramphenicol (C), streptomycin (S), tetracycline (TE), fosfomycin (FF) and erythromycin (E). Resistance to an antibiotic was confirmed using standard disk diffusion method. Break points to establish resistance were selected based on SFM recommendations for Enterobacteriaceae (Bonnet et al., 2010).
Among one hundred analyzed raw cow milk samples, seven were contaminated by Y. enterocolitica. Seventeen isolates were identified as Y. enterocolitica. Contaminated milk are from five regions: Amieur, Mansourah, Ouled Mimoun, Sebdou and Tlemcen. The greatest number of positive samples was obtained from Ouled Mimoun farms (42.85% with n=7). All the isolates were biotyped by biochemical tests and detection of the virulence genes confirmed their biotype as 1A. The results concerning phenotypic and biochemical characterics of Y. enterocolitica strains isolated from west Algerian raw milk are summarized in Table 1.
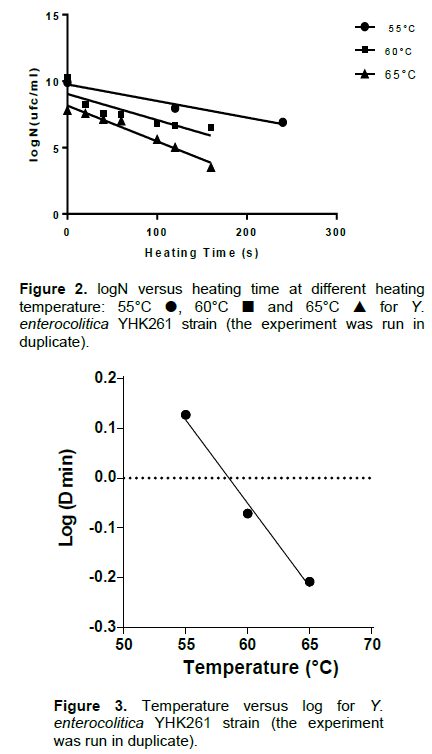
The thermal death curves at the three treatment temperatures and the D values are shown in Figure 2. The corresponding D55, D60 and D65 values were 1.34, 0.85 and 0.62 min, respectively. These values were deter-mined experimentally and Figure 3 shows how Z-value was determined. Z value was strongly elevated: 29.98°C.
The raw milk contamination frequency by Y. enterocolitica obtained in this study (7%) was higher than that reported in other studies, 1.6% in Iran (Soltan-Dallal et al., 2004) and 1% in Nigeria (Okeke and Okwori, 2014). Therefore, the assessment of Y. enterocolitica virulence indicators does not need to be restricted to the detection of plasmid-localized genes of virulence, but requires, at least, one chromosomal virulence-associated gene to be present (Ye et al., 2014). The ystB gene is present in all strains of biotype 1A, similar results were also found by Platt-Samoraj et al. (2006) and Jamali et al. (2015). Some researchers believe that these strains ystB+ are pathogenic to humans and can cause local outbreaks (Singh and Virdi, 2004b, 2005). The study of Singh and Virdi (2004b) indicated that the ystB gene is widely distributed in Y. enterocolitica biotype 1A strains and production of Yersinia stable toxin Yst-b produced by biotype 1A strains is the major contributor to diarrhea. The presence of ystB gene was often associated with clinical cases and represents a risk that should not be ignored. Our results show that it is possible to detect pathogenic strains with these traits.
Although, there is very little thermal inactivation data published in the scientific literature for Y. enterocolitica, Pagán et al. (1999) have reported D55 values of 0.33 to 0.78 min and D59 values of 0.18 to 0.6 min in citrate phosphate buffer. These values are less than those reported in this paper (1.34 and 0.85 min, respectively). A second study by Bolton et al. (2013) reported D55, D60 and D65 values of 10.98, 2.53 and 0.60 min, respectively, it is noteworthy that the D65 is the same as that obtained. The discrepancies of values may be attributed to the strain variation (Bhagat and Virdi, 2009). This study suggests that minor changes in the temperature of the milk treatment will greatly influence the survival of Y. enterocolitica and that mild temperatures are sufficient for the elimination of this microorganism as compared to others such as Mycobacterium avium subsp. paratuberculosis, Coxiella burnetii and bacterial spores (Pearce et al., 2012). Augmenting the temperature from 55 to 60°C would reduce the D values 1.6 fold, a time-temperature combination of 0.85 min at 60°C is required to achieve one log reduction in Y. enterocolitica, the equivalent time at 65°C was 0.62 min.
This study provides DT and Z values required to eliminate Y. enterocolitica and reduce the microbiological risk related to this microorganism without harming the organoleptic and nutritional quality of milk. Outbreaks caused by Y. enterocolitica strains have been reported after consumption of pasteurized milk (Ackers et al., 2000) however some heat resistance studies indicate that these strains are unable to survive to the pasteurization suggesting that their presence in pasteurized milk is either due to post-pasteurization contamination or under-processing (Greenwood et al., 1990). Y. enterocolitica can multiply at temperatures as low as 4°C (Bottone et al., 1999). Its presence in pasteurized milk, which is generally stored at refrigeration temperatures at the dairy, in the retail chain and at home, may have public health significance.
In Algeria, there is very little research related to food contamination by Y. enterocolitica. There are no reports of its presence in raw cow’s milk, pasteurized milk, or milk derivatives. Notably, it is usual in this country to prepare dairy products from unpasteurized milk, although this food can be a vehicle of pathogens to humans. This work shows the potential of public health risk in Algeria regarding infections transmitted by raw cow’s milk. Therefore, consumers are adviced to mild heat their milk before consumption to avoid the pathogenic Y. enterocolitica risk in raw milk or even in pasteurized milk.
Antibiotic susceptibility is of great importance. The results shown in Table 2 indicate that all the tested Y. enterocolitica strains (17/17) were susceptible to 13 antibiotics: Amikacin, Chloramphenicol, Ciprofloxacin, Gentamicin, Kanamycin, Naldixic acid, Streptomycin, Fosfomycin, Tetracyclin, Impinem, Piperacillin, Tobramycin and Trimethoprim/Sulfamethozol. Similar patterns of susceptibility were observed among strains isolated from pig tonsils in Switzerland, southern Germany as well as in human strains (Fredriksson-Ahomaa et al., 2009; Bucher et al., 2008). Piperacillin is a representative of ureidopenicillins with a broad scope of antibacterial activity. In the studies of Kot et al. (2008), 77.8% of Y. enterocolitica strains were found to belong to biotype 1A.
In veterinary medicine, treatment with antibiotics of penicillin group such as amoxicillin/clavulanic acid, ticarcillin/clavulanic acid, oxacillin, amoxicillin, ampicillin and piperacillin is frequent. The literature cites assessments of Y. enterocolitica susceptibility to Amoxicillin, as well as Amoxicillin/Clavulanic acid. In this study, resistance to Ampicillin was shown in all the 17 isolates. Ampicillin showed strong bactericidal activity towards a wide range of microorganisms; however, Y. enterocolitica strains are in general resistant to this antibiotic. Ampicillin resistance due to production of β-lactamases is well described in the literature (Bucher et al., 2008). We found no susceptible strains and only 1.94% of strains to be intermediately susceptible to Ampicillin. Singh and Virdi (2004a) found 100% of strains to be resistant, which is also confirmed by results from other authors (Rastawicki et al., 2000). The combination of ampicillin with clavulanic acid significantly broadens the scope of activity and increases the percentage of susceptible strains. In the present study, all strains were fully resistant to amoxicillin/clavulanic acid and ticarcillin/clavulanic acid. The study of Singh and Virdi (2004a) demonstrated that only 2.5% of Y. enterocolitica strains were intermediately susceptible, while the remaining strains were fully resistant to Amoxicillin/Clavulanic acid.
Fifteen strains of 17 (88.24%) are resistant to 1st generation Cephalosporins. This antibiotic group including Cefazolin, are active against Gram-positive and Gram-negative bacteria, while several studies proved that 90% of strains belonging to the Enterobacteriaceae family are resistant to Cefazolin (Fredriksson-Ahomaa et al., 2007; Bucher et al, 2008; Bhaduri et al., 2009). Kot et al. (2008) demonstrated that about 90% of biotype 1A Y. enterocolitica were susceptible to Cefazolin; these results are not in accordance with our data. Six (35.29%) strains are resistant to 2nd generation cephalosporins including cefoxitin and 6 (35.29%) strains are intermediately sensitive. The 2nd generation Cephalosporins have a stronger activity against Gram-negative than Gram-positive bacteria. Singh and Virdi (2004a) have demonstrated that 41.3 and 37.5%, of Y. enterocolitica strains are susceptible and 52.5 and 50% intermediately susceptible, respectively. The third generation Cephalosporins of which Ceftriaxone, Ceftazidime and Cefotaxime were included in earlier studies demonstrated bactericidal activity mainly against Staphylococcus sp. and Streptococcus sp., but also against Enterobacteriaceae, Haemophilus influenzae, Borrelia sp., and Pasteurella sp.
Y. enterocolitica are categorized as microorganisms susceptible to the 3rd generation Cephalosporins. In this study, 16, 10 and 8 (94.12, 58.82 and 47.06%) strains show resistance against Ceftriaxon, Ceftazidim and Cefotaxim, respectively. Singh and Virdi (2004a) did not find biotype 1A Y. enterocolitica strains resistant to the 3rd generation Cephalosporins. The studies of the Polish clinical strains of Y. enterocolitica, serotype O:3, have demonstrated full susceptibility of the strains to the 3rd generation Cephalosporins (Rastawicki et al., 2000). It may be concluded that in this study, the strains of Y. enterocolitica isolated from milk varied greatly in terms of their in vitro susceptibility to β-lactam antibiotics. Y. enterocolitica strains were found to be relatively highly resistant to Cephalosporins and most Penicillins. It is deemed necessary to systematically monitor the Y. enterocolitica strains susceptibility to antibiotics.
Due to Y. enterocolitica wide spread particularly in dairy product, inappropriate antibiotic treatment and prophylaxis, as well as antibiotics overuse in human and veterinary medicine, may lead to the development of resistant strains to one or several groups of antibiotics (Vose et al., 2001). Bacteria have wide mechanisms to develop antibiotic resistance, therefore it is important to systematically assess their susceptibility to individual antibiotics, thus enabling selection of an optimal treatment and preventing drug resistance spread among bacteria (Caprioli et al., 2000; Fredriksson-Ahomaa et al, 2009). The improper use of antibiotics in the developing countries like Algeria may be the main cause of high resistance rate in local Yersinia isolates.
Most importantly, preventive measures such as reasonable antibiotherapy must be adopted to avoid increasing resistance to antibiotics of Y. enterocolitica. Moreover using antibiotics as growth promoters must be prohibited.
This study provides data on the occurrence of Y. enterocolitica in raw milk in Western Algeria and their resistance ability to antibiotics. The risk due to the presence of Y. enterocolitica in raw milk is not insignificant. It highlights the role of raw milk and dairy derivatives as a transmission vehicle of potentially pathogenic Y. enterocolitica strains that can transmit antibiotic resistance to the intestinal flora. However, Y. enterocolitica is easily eliminated by heat treatment at mild temperatures of about 65°C.
The authors have not declared any conflict of interests.
REFERENCES
|
Ackers ML, Schoenfeld S, Markman J, Smith MG, Nicholson MA, DeWitt W, Slutsker L (2000). An outbreak of Yersinia enterocolitica O:8 infections associated with pasteurized milk. J. Infect. Dis. 181(5):1834-1837.
crossref
|
|
|
|
AFNOR (2003). Norme NF EN ISO 10273: Microbiologie des aliments : méthode horizontale pour la recherche de Yersinia enterocolitica présumées pathogènes. ISO 10273:2003.
|
|
|
|
Bari ML, Hossain MA, Isshiki K, Ukuku D (2011). Behavior of Yersinia enterocolitica in Foods. J. Pathog. 2011(13).
|
|
|
|
Bernardino-Varo L, Qui-ones-Ramírez EI, Fernández FJ, Vázquez-Salinas C (2013). Prevalence of Yersinia enterocolitica in raw cow's milk collected from stables of Mexico City. J. Food. Prot. 76(4):694-698.
crossref
|
|
|
|
Bhaduri S, Wesley I, Richards H, Draughon A, Wallace M (2009). Clonality and antibiotic susceptibility of Yersinia enterocolitica isolated from US market weight hogs. Foodborne. Pathog. Dis. 6(3):351-356.
crossref
|
|
|
|
Bhagat N, Virdi JS (2010). The Enigma of Yersinia enterocolitica biotype 1A. Crit. Rev. Microbiol. 1-15.
|
|
|
|
Bhagat N, Virdi JS (2009). Molecular and biochemical characterization of urease and survival of Yersinia enterocolitica biotype 1A in acidic pH in vitro. BMC. Microbiol. 9(1):262.
crossref
|
|
|
|
Bolton DJ, Ivory C, McDowell D (2013). Thermal inactivation of Yersinia enterocolitica in pork slaughter plant scald tank water. Meat Sci. 95(3): 668-671.
crossref
|
|
|
|
Bonnet R, Cavallo JD, Chardon H, Chidiac C, Courvalin P, Drugeon H, Dubreuil L, Guery B (2010). Comité de l'Antibiogramme de la Société Française de Microbiologie. CA. SFM. (2010):19-40.
|
|
|
|
Bottone EJ (1999). Yersinia enterocolitica: overview and epidemiologic correlates. Microbes. Infect. 1(4):323-333.
crossref
|
|
|
|
Bucher M, Meyer C, Grötzbach B, Wacheck S, Stolle A, Fredriksson- Ahomaa M (2008). Epidemiological data on pathogenic Yersinia enterocolitica in Southern Germany during 2000-2006. Foodborne. Pathog. Dis. 5(3):273-280.
crossref
|
|
|
|
Caprioli A, Busani L, Martel JL, Helmuth R (2000). Monitoring of antibiotic resistance in bacteria of animal origin: epidemiological and microbiological methodologies. Int. J. Antimicrob. Agents. 14(4):295-301.
crossref
|
|
|
|
Darwish SF, Asfour HA, Allam HA (2015). Incidence of Yersinia enterocolitica and Yersinia pseudotuberculosis in raw milk samples of different animal species using conventional and molecular methods. A. J. V. S. 44(1):174-185.
crossref
|
|
|
|
European Food Safety Authority (EFSA) (2011). The community summary report on trends and sources of zoonoses, zoonotic agents and foodborne outbreaks in the European Union in 2009. EFSA. J. 9(3):2090.
|
|
|
|
Falcão JP, Falcão DP, Pitondo-Silva A, Malaspina AC, Brocchi M (2006). Molecular typing and virulence markers of Yersinia enterocolitica strains from human, animal and food origins isolated between 1968 and 2000 in Brazil. J. Med. Microbiol. 55(11):1539-1548.
crossref
|
|
|
|
Fredriksson-Ahomaa M, Stolle A, Stephan R (2007). Prevalence of pathogenic Yersinia enterocolitica in pigs slaughtered at a Swiss abattoir. Int. J. Food. Microbiol. 119(3):207-212.
crossref
|
|
|
|
Fredriksson-Ahomaa M, Wacheck S, Koenig M, Stolle A, Stephan, R (2009). Prevalence of pathogenic Yersinia enterocolitica and Yersinia pseudotuberculosis in wild boars in Switzerland. Int. J. Food. Microbiol. 135(3):199-202.
crossref
|
|
|
|
Greenwood MH, Hooper WL, Rodhouse JC (1990). The source of Yersinia spp. in pasteurized milk: an investigation at a dairy. Epidemiol. Infect. 104(03):351-360.
crossref
|
|
|
|
Guven A, Sezer C, Aydin BD, Oral NB, Vatansever L (2010). Incidence and pathogenicity of Yersinia enterocolitica isolates from foods in Turkey. Kafkas Univ Vet Fak Derg. 16:107-112.
|
|
|
|
Huovinen E, Sihvonen LM, Virtanen MJ, Haukka K, Siitonen A, Kuusi, M (2010). Symptoms and sources of Yersinia enterocolitica-infection: a case-control study. BMC. Infect. Dis. 10(1):122.
crossref
|
|
|
|
ITELV (Institut technique des élevages en Algérie) (2012). Info élevage, bulletin trimestriel n°6. 2012. view
|
|
|
|
Jamali H, Paydar M, Radmehr B, Ismail S (2015). Prevalence, characterization, and antimicrobial resistance of Yersinia species and Yersinia enterocolitica isolated from raw milk in farm bulk tanks. J. Dairy. Sci. 98(2):798-803.
crossref
|
|
|
|
Jayarao BM, Henning DR (2001). Prevalence of foodborne pathogens in bulk tank milk. J. Dairy. Sci. 84(10):2157-2162.
crossref
|
|
|
|
Kot B, Jakubczak A, Piechota M (2008). Isolation and characterization of Yersinia enterocolitica rods from pig tonsils. Med. Weter. 64(3):283-287.
|
|
|
|
Lambertz ST, Danielsson-Tham ML (2005). Identification and characterization of pathogenic Yersinia enterocolitica isolates by PCR and pulsed-field gel electrophoresis. Appl. Environ. Microb. 71(7):3674-3681.
crossref
|
|
|
|
Mathew AG, Cissell R, Liamthong S (2007). Antibiotic resistance in bacteria associated with food animals: a United States perspective of livestock production. Foodborne Pathog. Dis. 4(2):115-133.
crossref
|
|
|
|
Okeke OFI, Okwori AEJ (2014). Occurrence of pathogenic Yersinia species in locally fermented cow milk (nono) in jos Nigeria. N. J. B. 23.
|
|
|
|
Pagán R, Ma-as P, Raso J, Trepat, FJS (1999). Heat resistance of Yersinia enterocolitica grown at different temperatures and heated in different media. Int. J. Food. Microbiol. 47(1):59-66.
crossref
|
|
|
|
Pearce LE, Smythe BW, Crawford RA, Oakley E, Hathaway SC, Shepherd, JM (2012). Pasteurization of milk: the heat inactivation kinetics of milk-borne dairy pathogens under commercial-type conditions of turbulent flow. J. Dairy. Sci. 95(1):20-35.
crossref
|
|
|
|
Perkowska K, Platt-Samoraj A, Bancerz-Kisiel A, Szweda W (2011). Susceptibility of Polish Yersinia enterocolitica strains isolated from pigs to 12 β- lactam antibiotics. Bull Vet Inst Pulawy. 55:397-402.
|
|
|
|
Plattâ€Samoraj A, Ugorski M, Szweda W, Szczerbaâ€Turek A, Wojciech K, ProcajÅ‚o Z (2006). Analysis of the presence of ail, ystA and ystB genes in Yersinia enterocolitica strains isolated from aborting sows and aborted fetuses. J. Vet. Med. 53(7):341-346.
crossref
|
|
|
|
Rahimi E, Sepehri S, Dehkordi FS, Shaygan S, Momtaz H (2014). Prevalence of Yersinia Species in Traditional and Commercial Dairy Products in Isfahan Province, Iran. Jundishapur. J. Microbiol. 7(4).
crossref
|
|
|
|
Rastawicki W, Gierczyński R, Jagielski M, Kałużewski S, Jeljaszewicz J (2000). Susceptibility of Polish clinical strains of Yersinia enterocolitica serotype O3 to antibiotics. Int. J. Antimicrob. Agents. 13(4):297-300.
crossref
|
|
|
|
Singh I, Virdi JS (2004a). In vitro susceptibilities of Yersinia enterocolitica biotype 1A. World. J. Microbiol. Biotechnol. 20:329-331.
crossref
|
|
|
|
Singh I, Virdi JS (2004b). Production of Yersinia stable toxin (YST) and distribution of yst genes in biotype 1A strains of Yersinia enterocolitica. Japan. J. Infect. Dis. 53(11):1065-1068.
crossref
|
|
|
|
Singh I, Virdi JS (2005). Interaction of Yersinia enterocolitica biotype 1A strains of diverse origin with cultured cells in vitro. Jpn. J. Infect. Dis. 58(1):31-33.
|
|
|
|
Soltan-Dallal MM, Tabarraie A, Moez Ardalan K (2004). Comparison of four methods for isolation of Yersinia enterocolitica from raw and pasteurized milk from Northern Iran. Int. J. Food. Microbiol. 94(1):87-91.
crossref
|
|
|
|
Subha B, Ramakrishnan D, Suganthi V (2009). Antimicrobial resistance pattern of selected Yersinia enterocolitica isolated from raw cow milk and pork samples of Namakkal District, Tamilnadu, South India. I. J. E. R. 3(3):169-177.
|
|
|
|
Tennant SM, Grant TH, Robinsâ€Browne RM (2003). Pathogenicity of Yersinia enterocolitica biotype 1A. FEMS. Immunol. Med. Microbiol. 38(2):127-137.
crossref
|
|
|
|
Tudor L, Ţogoe I, Ceauşi C, Mitrănescu E, Goncearov M, Rotaru E (2008). The optimization of Yersinia enterocolitica isolation and identification from food products. Lucrari. Stiintifice. USAMV.41:993-1002.
|
|
|
|
Vose D, Acar J, Anthony F, Franklin A, Gupta R, Nicholls T, Tamura Y, Thompson S, Threlfall EJ, Van Vuuren M, White DG, Costarrica ML (2001). Antimicrobial resistance: risk analysis methodology for the potential impact on public health of antimicrobial resistant bacteria of animal origin. Rev. Sci. Tech. OIE. 20(3):811-827.
|
|
|
|
Wang X, Cui Z, Wang H, Tang L, Yang J, Gu L, Jin D, Luo L, Qiu H, Xiao Y, Xiong H (2010). Pathogenic strains of Yersinia enterocolitica isolated from domestic dogs (Canisfamiliaris) belonging to farmers are of the same subtype as pathogenic Y. enterocolitica strains isolated from humans and may be a source of human infection in Jiangsu Province, China. J. Clin. Microbiol. 48(5):1604-1610.
crossref
|
|
|
|
Ye YW, Ling N, Han YJ, Wu QP. (2014). Detection and prevalence of pathogenic Yersinia enterocolitica in refrigerated and frozen dairy products by duplex PCR and dot hybridization targeting the virF and ail genes. J. Dairy. Sci. 97(11): 6785-6791.
crossref
|