ABSTRACT
Soil treatment incubation method was carried out to analyze the monocrotophos (MCP) biodegradation using three selected bacterial isolates namely Pseudomonas stutzeri (KY287931), Bacillus licheniforms (KY287928), and Bacillus sonorensis (KY287930). After 45 days of incubation of pesticide exposed to soil samples, the MCP biodegradation by individual cultures and consortium culture was analyzed and conformed by gas chromatography–mass spectrometry (GC-MS) method. The three isolates individually degrade toxic MCP into non-toxic intermediate while the consortium cultures completely degrade the MCP into water-soluble compounds. PCR-based RFLP analysis of 16S rRNA was performed. Pseudomonas and Bacillus species have been phylogenetically analyzed by using 16S rRNA gene sequences. The isolated bacterial consortium culture proved a potential agent for MCP biodegradation.
Key words: Soil, MCP-monocrotophos, bacterial isolate, gas chromatography–mass spectrometry (GC-MS).
Use of microorganisms for properly detoxifying, degrading and eradication of toxic compounds from contaminated soil and water has emerged as the best technique to clean up contaminated environments (Strong and Burgess, 2008). Monocrotophos (MCP) is still widely used in India for the protection of cash crops such as cotton, sugarcane, tobacco, maize, groundnut, soybeans, rice, and vegetables (Acharya et al., 2015). Microorganism plays a major role in agriculture as they increase the exchange of plant nutrients and make chemical fertilizers usage minimal as much as possible.
Beneficial plant-microbe interactions in the rhizosphere can improve the soil fertility (Dastager et al., 2011). Microbes plays a crucial role for changeover of phosphorous in water and sediments and the phosphate ions are reported strongly, absorbed by sediments with a high content of slit and clay (Chen et al., 2015). In order to accomplish an effective degradation, the microorganisms were isolated from agricultural fields which were previously exposed to the pesticides (Jayanthi et al., 2014). Organophosphate hydrolase (opdA gene) wrapped up in starting biodegradation of monocrotophos in KPA-1 was quantitatively shown, which fundamentally indicates cytosolic enzyme; a catabolic pathway for MCP degradation has been proposed (Bhadbhade et al., 2002). Numerous problems associated with pesticides application are their possible persistence in the ecosystem and therefore, their possible integration into the food chain affects ecosystems and human population (Hill et al., 2003). In the recent years soil quality deterioration has caused a serious concern to our planet. Pesticide contaminations of soil pose a grave danger to human health as well as to the natural environment. The methods procurable today (physical or chemical) are on the other hand costly or incomplete. Bioremediation contribute a superior process or tools for the efficient detoxification of pesticides from the soil. Bioremediation is environmentally healthy, cost effective and has efficient method (Baba et al., 2016).
The pathway is initiated by the conversion of dimethyl (E)-1-methyl-2-(methylcarbamoyl) vinyl phosphate and then to N-methylhydroxyl monocrotophos dimethyl phosphate; both steps are catalyzed dependently by dimethylphosphate. N-methylacetoacetamide is transformed to 3-hydroxy-N-methylbutyramide, and methyl phosphate is converted to phosphoric acid. Microorganisms contribute a great percentage in the conversion of phosphorous in water and the phosphate ions are actively taken by ground with major portion of clay and slit (Seema et al., 2013).
Pesticides can be degraded by microbial, chemical and photo degradation processes in the environment. Nonetheless, microbial degradation is considered as the major determining factor of the organophosphrus fate in the environment and is often the main process of pesticide degradation in soils, representing the safest, least disruptive and most cost-effective treatment method (Zrostlikova et al., 2003; Singh et al., 2004). Several researchers investigated the capabilities of bacteria to degrade MCP (Singh et al., 2009). All of these enzymes are found to induce the degradation of organophosphate compounds.
The aim of this work is to study the impact of MCP on soil microorganisms. Twenty-five safe bacterial species were isolated, purified and identified from soil with history pesticide application, then used to estimate their effect on the biodegradation rate of tested organophosphrus insecticides, to give a primitive indication about organophosphrus insecticides biodegradation.
Bacterial samples
Three promising MCP degrading bacterial isolates-from our previous work (Buvaneswari et al., 2017), namely Pseudomonas stutzeri KY287931, Bacillus licheniformis KY287928, and Bacillus sonorensis KY287930 were selected for the present study.
Soil treatment incubation
The organophosphorus pesticide monocrotophos used at concentration of 1000 ppm was mixed with 250 g of soil already dried and sterilized. The autoclaved soil and water was used as control and the treatments made were as follows:
Control- Soil + Pesticide (1000ppm)
Treatment 1- Soil + Pesticide (1000ppm) + P. stutzeri (100ml)
Treatment 2- Soli + Pesticide (1000ppm) + Bacillus licheniforms (100ml)
Treatment 3- Soil + Pesticide (1000ppm) + B. sonorensis (100ml)
Treatment 4- Soil + Pesticide (1000ppm) +Consortium (100ml)
100 g of soil was added in 100 ml of sterilized water for the preparation of soil culture. Individual treatments of soil in triplicate were flooded with inoculum to keep them moist during incubation period. These treatments were kept at incubator (28 to 30°C) under darkness for 45 days. Two sets of flasks were prepared with the pesticides at 1000 ppm. After the completion of incubation, period flasks were taken out accordingly for extraction and analysis (Wasim et al., 2009).
Extraction and analysis
25 g of treated soil (samples) was air dried for extraction and homogenized with 0.5 g charcoal (activated) for 4 h at 120°C. Then added 1.0 g Florisil (activated for 4 h at 650°C) and 5 drops of 25% NH3H2O solution placed over a 2.5 cm layer of anhydrous sodium sulphate (Parthiban et al., 2015) (analytical grade) in a glass column with 34 cm length and 2.5 cm dia.
Extraction was done, using a solution of n-hexane (distilled) and acetone (distilled) in a ratio of 2:1 by the method described by Wasim Aktar et al. (2009). Eluted material was collected in a 250 ml conical flask (Pyrex) and later evaporated on rotary evaporator to almost dryness dissolved in 20 to 50 µl quantity n-hexane in small glass vials for Gc-ms determination.
Analysis of liquid samples on the Agilent GC-MS
Gas chromatographic determination
The samples were analyzed in GCMS (Gas Chromatograph- Mass Spectrophotometer) (5975c: Insert MSD with Triple Axis Detector). The limit of detection (LOD) of OCPs was determined, the blank. Before analysis, standards were run to check for the column performance, peak height and resolution. The extract was analyzed on GC (equipped with flame ionization detector) with the parameters as column temperature 230°C, injector temperature 250°C, detector temperature 300°C, hydrogen gas flow 4.5 ml/min and air flow 175 ml/min.
The parameter of the glass column was 1.5 % OV-17 + 1.75 % OV-210 Chrom W-HP 80/100 mesh 2 meters x ¼” x 2 mm ID (internal diameter). The retention time was 1.6 min, detection limit was 1µg and recovery percentage was 100%. An Agilent Gas Chromatography system 5975c with Mass Selective Detector 5973 was used. If the retention time, qualifying ions (molecular weights) and response (area on the graph giving the abundance corresponding to the concentration) values were in the correct range and the Q value (accuracy estimation from the machine in percentage) was high then, the result was accepted and process (Mishra et al., 2017; Gilani et al., 2010).
Molecular characterization of MCP degrading bacterial isolates
Restriction fragment length polymorphism (RFLP) analysis
Extraction of genomic DNA from MCP degrading bacterial strains: The isolates were used to inoculate in nutrient broth (Hi-media) and incubated overnight at 37°C and 200 rpm. The resulting bacterial suspension was pelleted at 10,000 rpm for 5 min and the genomic DNA was extracted from the samples using phenol-chloroform extraction technique. The buffer in the micro centrifuge tube contains the DNA. The DNA samples were stored at -20°C until further use (Lori et al., 2005; John, 2014).
Purification of 16S rRNA PCR product
The partial 16S rRNA gene fragments were amplified from the cultures with PCR conditions as follows: the PCR products were precipitated with 9 μl of 3 M sodium acetate (pH 5.2) and 60 μl of isopropanol and resuspended in 50 μl of distilled water. Extraction of the PCR products with chloroform was carried out (Akhter et al., 2013).
Electrophoresis and enzymatic digestion of amplified DNA
Purified DNA was digested for 2 h in 10 μl volumes with restriction endonucleases. The following three restriction enzymes were used: ALU1, SAU3A1and HAE. Triple digestion with SAU3A1 and ALU1 and HAE was performed as follows: DNA was first digested for 1 h at 25°C with ALU1.
Following the addition of buffer recommended by the manufacturer, SAU3A1and HAE were added and the reaction mixture was incubated for 1 h at 37°C (Daniel et al., 2009). The resulting DNA fragments were electrophoresed in 1.5% (wt/vol) agarose gels in Trisacetate buffer (0.04 M Tris-acetate, 0.001 M EDTA; pH 8.3) at 100 V for 30 min. The gels were stained with ethidium bromide and DNA fragments were visualized by UV transillumination and photographed. Strains were categorized according to their distinct RFLPs (Henrik, 2012).
Phylogenetic analysis data
To assess the phylogeny of the isolates, a BLASTN analysis (16S rRNA sequence) was carried out through GenBank (Liu et al., 2001; Castresana et al., 2000; Dereeper et al., 2008).
Construction of phylogenetic trees
The computer assisted by data analysis was performed for RFLP profiles. Two numbers of alternative trees were constructed. Negative branch lengths were avoided or minimized in the best trees selected. Farris trees were constructed according to Archibald (2002) and UPGMA trees as described by Allman et al. (2017).
In our previous study, a total of 121 bacteria isolates belong to three genera, capable of MCP degradation with particular concentration which were studied and conformed by FTIR (Gilani et al., 2010). All of the pure bacterial isolates were sequentially subcultures on mineral salt medium.
Similarly, several species of bacteria were isolated from different environment which degrade organophosphorus compound in laboratory cultures and soil (Buvaneswari et al., 2017). Monocrotophos degradation in soil treatment incubation method using selected bacterial isolates was analyzed and conformed by GC-MS method. After forty five days of inoculation with the selected bacterial species, the consortium is compared with uninoculated samples. The metabolic products of MCP degradation by strains P. stutzeri (KY287931), B. licheniforms (KY287928), and B. sonorensis (KY287930) filtrates were extracted and confirmed by GC-MS. Sasano et al. (2000) proposed the analysis of pesticides in water samples using the GC/MS technique (Singh et al., 2006).
Gas chromatographic mass-spectrum is generated by monocrotophos in control culture media (Figures 1 and 2). The control sample compared to NIST library identification program have a phosphoric acid, dimethyl1-methyl-3-(metylamino)-3-oxo-1-propenyl ester group compound (C7H14NO5P). The degradation of organophosphate pesticide by microorganisms appears to be identical, where OPH or PTE catalyzes the first step of the degradation (Sasano et al., 2000). The peaks at 11.6 min indicate that 2-hydroxy-4-hydroxylaminopyrimidine is one of the most important compounds in monocrotophos and peaks at 12.86 min indicate phosphoric acid and dimethyl 1-methylpropenyl ester. Each of the peaks was identified on the basis of its mass spectra and the NIST library identification program. These compounds strongly evidence the presence of MCP in the extracted control samples. Anizio et al. (2009), proposed a method of analysis for six pesticides (imazethapyr, imazaquin, metsulfuron-methyl, bentazone, chlorimuron-ethyl and tebuconazole), by GC with mass spectrometer detector (Anizio et al., 2009).
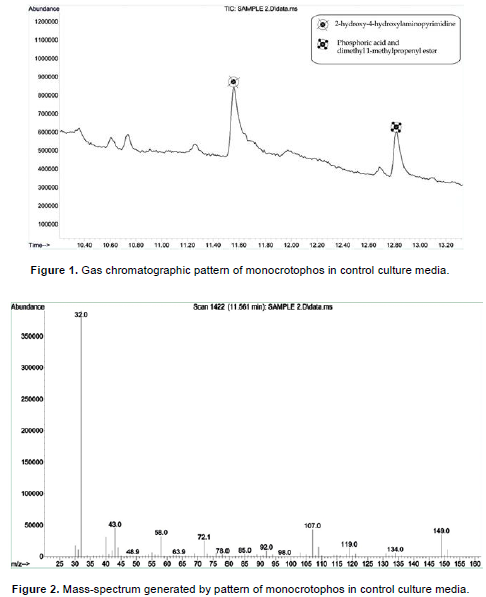
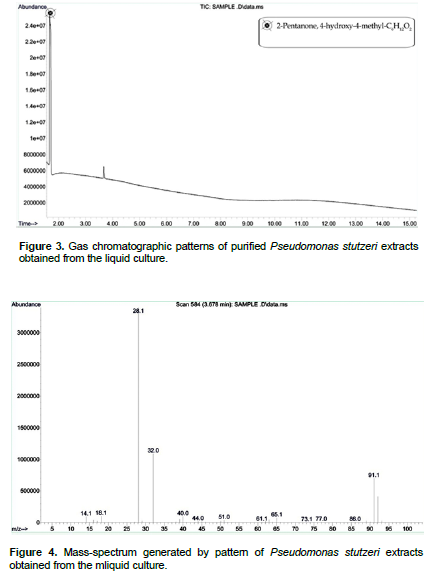
After 45th day of incubation, the gas chromatographic mass-spectrum generated by KY287931 (P. stutzeri) culture media was analyzed (Figures 3 and 4). The peak at retention time 11.6 min (2-hydroxy-4-hydroxylaminopyrimidine) and 12.86 min (phosphoric acid and dimethyl 1-methylpropenyl ester) corresponded to control sample, this peak disappeared concomitantly with the formation of another new peak with a retention time of around 1.668 min. The new peak with a retention time of around 1.668 min indicate 2-pentanone, 4-hydroxy-4-methyl-C6H12O2.The results demonstrated that P. stutzeri is a potent and easily acquired bioremediation agent that could be used to remove other pollutants from pesticide contaminated soils. A similar study was conducted by Aleem (2003), who investigated the determination of organochlorine pesticides in agricultural soil with special reference to γ-HCH degradation by Pseudomonas strains.
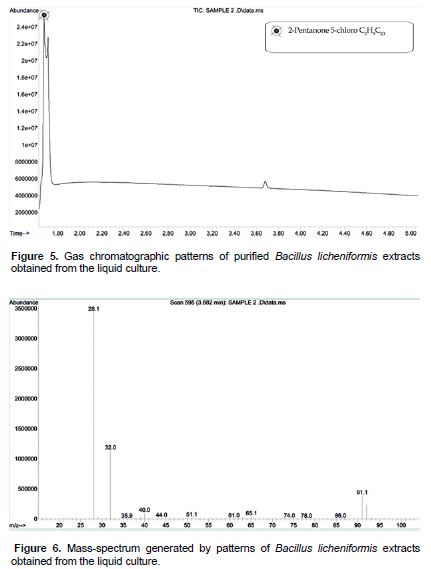
After 45th day of incubation, the gas chromatographic mass-spectrum generated by BAGN005 (B. licheniformis) culture media were analyzed (Figures 5 and 6). The peak at retention time of 11.6 min (2-hydroxy-4-hydroxylaminopyrimidine) and 12.86 min (phosphoric acid and dimethyl 1-methylpropenyl ester) corresponded to control sample. This peak disappeared concomitantly with the formation of another new peak through a retention time of around 1.676 min. Zhao et al. (2012) described the same method to study pesticides in vegetables. The graph shows peak with a retention time of around 1.676 min, which indicates the 2-pentanone 5-chloro C5H9ClO. This is one of the four stable isomers of butanediol. In biology, 1, 3-butanediol is used as a hypoglycemic agent.
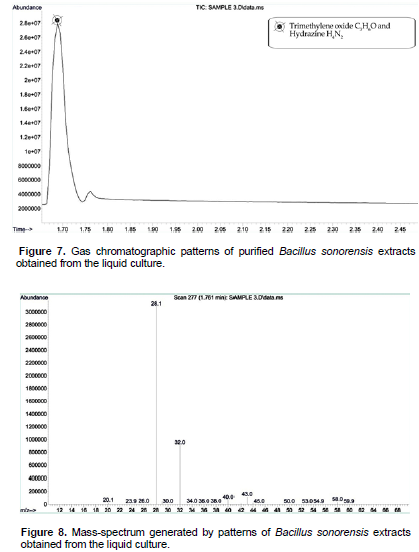
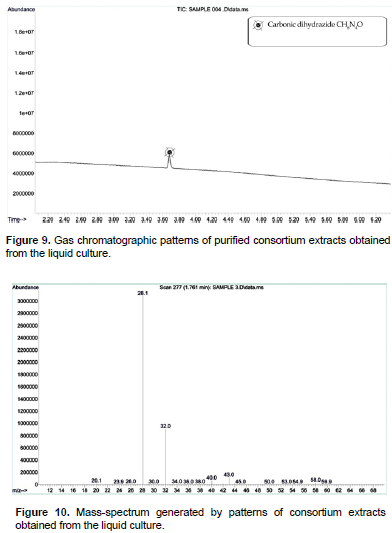
After 45th day of incubation, the gas chromatographic mass-spectrum generated by BKGN007 (B. sonorensis) culture media were analyzed (Figures 7 and 8). The peak at retention time of 11.6 min (2-hydroxy-4-hydroxylaminopyrimidine) and 12.86 min (phosphoric acid and dimethyl 1-methylpropenyl ester) corresponded to control sample. This peak disappeared concomitantly with the formation of another new peak through a retention time of around 1.495 min. The results demonstrated that B. sonorensis is a potent and easily acquired bioremediation agent that could be used to remove other pollutants from pesticide contaminated soils. The new peak with a retention time of around 1.495 min indicates the trimethylene oxide C3H6O and hydrazine H4N2. Hydrazine is a convenient reductant because the by-products are typically nitrogen gas and water. Thus, it is used as an antioxidant and an oxygen scavenger. After 45th day of incubation, the gas chromatographic mass-spectrum generated by mixed consortium culture media were analyzed (Figure 9 and 10). The peak at retention time of 11.6 min (2-hydroxy-4-hydroxylaminopyrimidine) and 12.86 min (phosphoric acid and dimethyl 1-methylpropenyl ester) corresponded to control sample, this peak disappeared concomitantly with the formation of another new peak through a retention time of around 3.60 min (Mishra et al., 2017; Gilani et al., 2010). The new peak with a retention time of around 3.60 min indicates the carbonic dihydrazide CH6N4O. The result presented good results for the microbial degradation of monocrotophos. Carbohydrazide is the chemical compound with the formula OC (N2H3)2. It is a white and water-soluble solid. It decomposes upon melting. A number of carbazides are known where one or more N-H groups are replaced by other substituents. The accumulation of the products was not found by the determination after incubation for 45 days. The degradation products were similar to those observed in previous studies (John et al., 2004; Horsfield et al., 2008; Singh et al., 2003). They occur widely in the drugs, herbicides, plant growth regulators, and dyestuffs. GC-MS analysis showed that consortium culture has more capacity to degrade the MCP than other isolated microorganisms. The ester metabolite is detected by GC-MS analysis; it is eco- friendly for soil environment. However, in the present study, degradation of organophosphorus pesticide was obtained and this could be the reasoned why bacterial strains consortium express the pesticide degrading enzymes (Singh et al., 2009).
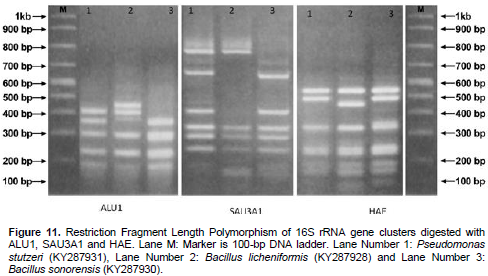
In the present study, total DNA was extracted according to the procedures described by M. A. Akhter and R. Laz, (2013). The isolated chromosomal DNA was 1500 bp which compared with 1kb + DNA marker (Figure 11). The RFLPs of 16S rRNA were determined using three restriction enzymes, ALU1, SAU3A1 and HAE. PCR-based RFLP analysis of 16S rRNAs was performed. The genetic diversity assessment by RFLP shows genetic similarity between the isolates which were varied (Figure 11). Pseudomonas and Bacillus species have been phylogenetically analyzed by using 16S rRNA gene sequences (Bhadbhade et al., 2002).
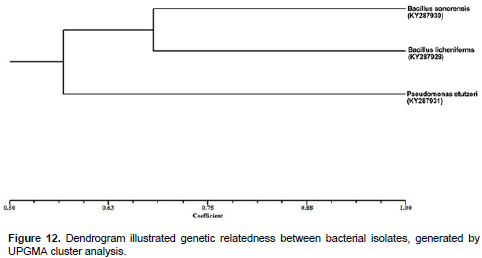
The distinct RFLP pattern of the 3 strains could be due to increased mutation under stress, facilitation adaptation of the strains to stressful environments and also due to the different types of pesticide contamination and varying age of contamination at the sites. Phylogenetic analysis was done on the basis of partial 16S rRNA sequences (600 nucleotides) of 3 isolates. The 3 strains were grouped into two clusters, the blast similarity search and phylogenetic analysis of 16S rRNA sequence which revealed that, the isolated bacterial strain are closely similar to Pseudomonas and Bacillus groups (Figure 12). Dendrogram analysis Illustrate the relationship between strains of P. stutzeri KY287931, B. licheniforms KY287928 and B. sonorensis KY287930 bacteria. Based on the results of physiological, biochemical and molecular analysis, the isolates were designated. At present, more than 20 isolates of MCP-degrading strains have been reported and yielded a variety of microorganisms.
With this increased awareness, research has recently focused to the fate of pesticides in soils and health risk due to their potential transfer and accumulation in plants. Our study reveals that isolated bacterial strains growing in contaminated sites are able to with-stand high levels of monocrotophos in the soil. The consortium culture shows increased degradation activity in the MCP contaminated soil as compared to individual cultures.
MCP was highly adsorbed by the studied isolates. The bacterial samples were identified and confirmed as P. stutzeri KY287931, B. licheniforms KY287928, and B. sonorensis KY287930 through Restriction Fragment Length Polymorphism of 16S rRNA gene clusters.
The authors have not declared any conflict of interests.
The authors are grateful to the Management and the Principal of J.J College of Arts & Science (A), Namunasamudram, Sivapuram, Pudukkottai, Tamilnadu, India for providing all the necessary facilities and encouragement. We would like to acknowledge the helpful and extensive recommendations of anonymous reviewers made to this paper. Dr. N. Thajuddin wish to acknowledge financial support from UGC-BSR One Time Grant (Ref.F.19-156/2015 (BSR) Oct.2015.
REFERENCES
|
Acharya KP, Shilpkar P, Shah MC, Chellapandi P (2015). Biodegradation of insecticide monocrotophos by Bacillus subtilis KPA-1, isolated from agriculture soils. Appl. Biochem. Biotechnol. 175(4):1789-804.
Crossref
|
|
|
|
Akhter MA, Laz R (2013). Isolation and Molecular Characterization of Pesticide (Fenitrothion) Resistant Bacteria from Agricultural Field, IOSR J. Pharm. (e)-ISSN:2250-3013.
|
|
|
|
Aleem A, Malik A (2003). Determination of organochlorine pesticides in agricultural soil with special reference to γ-HCH degradation by Pseudomonas strains. Biores. Technol. 88(1):41-46.
Crossref
|
|
|
|
Allman E, Degnan J, and Rhodes J (2017). Species tree inference from gene splits by unrooted star methods. IEEE/ACM Transactions on Computational Biology and Bioinformatics 99:1-1.
|
|
|
|
Anizio M, Faria Carol H, Collins and Isabel CSF, Jardim (2009). State-of-the-Art in Immobilized Polymer Stationary Phases for High-Performance Liquid Chromatography. J. Braz. Chem. Soc. 20(8):1385-1398.
Crossref
|
|
|
|
Archibald J, Roger A (2002). Gene conversion and the evolution of euryarchaeal chaperonins: a maximum likelihood– based method for detecting conflicting phylogenetic signals. J. Mol. Evol. 55:232-245.
Crossref
|
|
|
|
Baba Uqab, Syeed Mudasir, Ruqeya Nazir (2016).Review on Bioremediation of Pesticides. J. Bioremediat. Biodegrad.7:3.
|
|
|
|
Bhadbhade BJ, Sarnaik SS, Kanekar PP (2002). Biomineralization of an organophosphorus pesticide, Monocrotophos, by soil bacteria. J. Appl. Microbiol. 93:224-234.
Crossref
|
|
|
|
Buvaneswari G, Thenmozhi R, Nagasathya A and Thajuddin N (2017). Screening of Efficient Monocrotophos Degrading Bacterial Isolates from Paddy Field Soil of Sivagangai District, Tamil Nadu, India. J. Environ. Sci. Technol. ISSN 1994-7887.
|
|
|
|
Castresana J (2000). Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Molec. Biol. Evol. 17:540-552.
Crossref
|
|
|
|
Chen M, Xu P, Zeng G, Yang C, Huang D, and Zhang J (2015). Bioremediation of soils contaminated with polycyclic aromatic hydrocarbons, petroleum, pesticides, chlorophenols and heavy metals by composting, Biotech. Adv. 33:745-755.
Crossref
|
|
|
|
Daniel G Gibson, Lei Young, Ray-Yuan Chuang, Craig Venter J, Clyde A Hutchison and Hamilton O Smith (2009). Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6:343-345.
Crossref
|
|
|
|
Dastager SG, Deepa CK, and Pandey A (2011). Potential plant growth promoting activity of Serratia nematodiphila NII- 0928 on black pepper. World J. Microbiol. Biotechnol. 27:259-265.
Crossref
|
|
|
|
Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M, Claverie JM (2008). Phylogeny. fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36(suppl_2):W465-W469.
|
|
|
|
Gilani STS, Ageen M, Shah H and Raza S (2010). chlorpyrifos degradation in soil and its effect on soil microorganisms. J. Animal Plant Sci. Page:99-102. ISSN:1018-7081.
|
|
|
|
Henrik BR (2012). Restriction fragment length polymorphism analysis of PCR-amplified fragments (PCR-RFLP) and gel electrophoresis-valuable tool for genotyping and genetic fingerprinting. InGel Electrophoresis-Principles and Basics. InTech. pp.315-334, ISBN 978-953-51-0458-2.
|
|
|
|
Hill TCJ, Walsh KA, Harris JA, and Moffett BF (2003). Using ecological diversity measures with bacterial communities. FEMS Microbiol. 43:1-11.
Crossref
|
|
|
|
Horsfield A, Sallam MN, Drummond FA, Williams DJ, Schultz RJ (2008). Role of climatic factors on damage incidence by Dermolepida albohirtum (Coleoptera: Scarabaeidae), in Burdekin sugarcane fields, Australia. J. Econ. Entomol.101 (2):334-40.
Crossref
|
|
|
|
Jayanthi A, Sivagnanam S, Peter L (2014), Simultaneous degradation of organophosphorus and organochlorine pesticides by bacterial consortium. J. Taiwan Institute Chem. Eng., pp. 2590-2596.
|
|
|
|
John L, Fostera Bia R, KwanaTonyVancovb H (2004). Microbial degradation of the organophosphate pesticide. Ethion 240(1):49-53.
|
|
|
|
John TC (2014). Basic Molecular Protocols in Neuroscience: Tips, Tricks, and Pitfalls. Academic Press ISBN: 9780128014615.
|
|
|
|
Liu YH, Chung YC, Xiong Y (2001) Purification and characterization of adimethoate-degrading enzyme of Aspergillus niger ZHY256, isolated from sewage. Appl. Environ. Microbiol. 67:3746-3749.
Crossref
|
|
|
|
Lori S, Eggert Jesus E, Maldonado Robert , Fleischer C (2005). Nucleic acid isolation from ecological samples animal scat and other associated materials. Methods Enzymol. pp. 73-82.
|
|
|
|
Mishra A, Jamaluddin and Pandey AK (2017).A Review on Microbial Degradation of Organophosphorous Pesticide: Methyl Parathion, Austin J. Biotechnol. Bioeng. 4(1):1074.
|
|
|
|
Parthiban K, Vignesh V, Nilmini V, Thirumurugan R (2015). A statistical approach on optimization of exopolymeric substance production by Halomonas sp. S19 and its emulsification activity. Bioresour. Bioprocess. 2:48.
Crossref
|
|
|
|
Sasano R, Hamada T, Kurano M, Furuno (2000). On-line coupling of solid-phase extraction to gas chromatography with fast solvent vaporization and concentration in an open injector liner. Analysis of pesticides in aqueous samples. J. Chromatogr. 896:41- 49.
Crossref
|
|
|
|
Seema BS, Sayyed RZ, Trivedi MH, Gobi TA (2013). Phosphate solubilizing microbes: sustainable approach for managing phosphorus deficiency in agricultural soils. SpringerPlus. 2(1):587.
Crossref
|
|
|
|
Singh BK, Walker A (2006). Microbial degradation of organophosphorus compounds. FEMS Microbiol. Rev. 30:428-71.
Crossref
|
|
|
|
Singh BK, Walker A, Morgan AW, Wright DJ (2003). Effects of soil pH on the biodegradation of chlorpyrifos and isolation of a chlorpyrifos-degrading bacterium. Appl. Environ. Microbiol. 69:5198-5206.
Crossref
|
|
|
|
Singh P, Dhanjal S, Singh KV, Suri CR, Cameotra SS (2009). Monitoring atrazine uptake by a new fluorometric assay. Current Sci. 25:268-271.
|
|
|
|
Singh P, Suri CR, Cameotra SS (2004). Isolation of a member of Acinetobacter species involved in atrazine degradation. Biochemical and biophysical research communications. 317(3):697-702.
Crossref
|
|
|
|
Strong PJ, Burgess JE (2008). Treatment methods for wine-related and distillery wastewaters review. Bioremedy J. 12:70-87.
Crossref
|
|
|
|
Wasim Aktar Md, Dwaipayan Sengupta, and Ashim Chowdhury (2009). Impact of pesticides use in agriculture their benefits and hazards. Interdiscip Toxicol. 1-12.
Crossref
|
|
|
|
Zhao, Suli, Zhai, Andy (2012). A blind study of pesticides in vegetables by agilent bond elut quechers extraction kits and agilent 5975T LTM GC/MSD, Agilent Technologies, Inc. Publication number 5990-6323EN.
|
|
|
|
Zrostlikova J, Hajslova J, Cajka T (2003). Evaluation of two dimensional gas chromatography-time-of-flight mass spectrometry for the determination of multiple pesticide residues in fruit. J. Chromatogr. 1019:173-186.
Crossref
|