Full Length Research Paper
ABSTRACT
Poor effluent management is known to release antibiotic resistance genes and heavy metal resistance genes into streams. The objective of this study was to investigate the occurrence of antibiotics and heavy metals in hospital effluents and streams in Benin. The extraction of genomic DNA from multidrug-resistant bacterial strains isolated from stream and hospital effluents samples was performed according to the recommendations of the Quick-DNA TM miniprep kit (Zymo Research Corp, United States). Real-time PCR was used to identify twelve antibiotic and six heavy metal resistance genes. The results showed that sulII (77.77%), sulI (67.67%), and blaTEM-1 (44.44%) were the resistance genes to antibiotics, the most detected in gram-negative bacilli isolated from hospital effluents. Two genes, tetA (33.33%) and ermB (20%), were found in gram-positive cocci. zntA (57.57%), czcA (24.24%), and copA (22.22%) are the genes encoding resistance to heavy metals, most found in gram-negative bacilli, but zntA (20%) and czcA (10%) were both found in Staphylococcus aureus isolates. Concerning streams, sulII (38.23%), sulI (26.47%), and blaTEM-1 (23.53%) were detected in gram-negative bacilli. czcA (38.23%), zntA (35.29%), and copA (11.76%) are the genes encoding heavy metal resistance found in gram-negative stream bacilli. These results highlight the need for measures to be taken to ensure the integrity of natural resources and thereby preserve human, animal and environmental health.
Key words: Antibiotic and heavy metal resistance genes, hospital effluents and streams, Benin.
INTRODUCTION
Water is one of the most precious resources on earth. For humans, it is one of the basic needs used for food and other ancillary needs. It is therefore agreed that its availability in quantity and quality is essential for life on earth. Among these resources, streams represent one of the most important resources after the oceans. It is widely used for many human activities, such as fishing, agriculture, transportation, and many others. However, it is subject to numerous contaminations. This contamination can come from various sources related to human activities, such as industrial, agricultural, domestic discharges and hospital effluents (Adelowo et al., 2018; Kayembe et al., 2018; Zhou and Wang, 2019; Laffite et al., 2020; Yeh et al., 2020; Duan et al., 2021). The latter is one of the most important sources of contamination. Indeed, the contamination of streams can be of various natures that are chemical, microbiological, and sometimes radioactive (Pietryczuk et al., 2018; Carles et al., 2019; Mandaric et al., 2019; Yeh et al., 2020). Given that hospitals are environments par excellence where we witness the presence of large numbers of microbes of various kinds (bacteria, parasites, fungi, and viruses), these hospital effluents therefore contribute strongly to the microbiological contamination of streams (Verburg et al., 2019; Suzuki et al., 2020). In addition to microbes, the most important of which are bacteria, hospital effluents also contain antibiotic residues and resistance genes. Among the priority antibiotic-resistant pathogens, carbapenem-resistant Acinetobacter baumannii, carbapenem-resistant Pseudomonas aeruginosa, carbapenem-resistant and ESBL-producing Enterobacteriaceae are at the top of the WHO list and represent the greatest threat to human health (Sheu et al., 2019). In addition, Colistin regained global interest as a consequence of the rising prevalence of multidrug-resistant Gram-negative Enterobacteriaceae. In parallel, colistin resistant bacteria emerged in response to the unregulated and increased use of this antibiotic, which is a last resort drug due to failure of carbapenems, has possibly contributed to the development and spread of resistance to colistin among Enterobacteriaceae (Gogry and Siddiqui, 2019; Gogry et al., 2021, 2022). Indeed, many studies have shown the presence of these priority resistant pathogens in stream and hospital effluents (Adelowo et al., 2018; Bartley et al., 2019; Posada-Perlaza et al., 2019; Niest?pski et al., 2020; Suzuki et al., 2020). Even if other activities, such as livestock farming and domestic water, can bring these elements into watercourses, hospital effluents are suspected to be one of the main sources (Adelowo et al., 2018).
Bacteria are known to be the most prevalent in the hospital environment and in these effluents (Giannakis et al., 2017; Mittelman and Jones, 2018). These bacteria are most often multidrug-resistant to antibiotics due to the presence of antibiotics in the environment and the strong transmission of resistance genes (Giannakis et al., 2017; Mittelman and Jones, 2018; Laffite et al., 2020). Additionally, the presence of heavy metals in the medical environment exerts pressure favoring the selection of opportunistic pathogens resistant to antibiotics. Heavy metal resistance (HMR) associated with antibiotic resistance (AR) in hospital effluents makes them potentially dangerous (Chen et al., 2019; Dahanayake et al., 2019; Turner et al., 2020). Moreover, several studies have shown the presence of antibiotic and heavy metal resistance genes (ARGs and HMRGs) in wastewater, sewage sludge, river water, and Black sea (Sabatino et al., 2020; Hubeny et al., 2021; Martin et al., 2021). Other studies have established the correlation between these two types of resistance (Di Cesare et al., 2016; Yuan et al., 2018; Ohore et al., 2019). This is why it is advisable to have a water treatment and purification system in every hospital or city. Even if industrialized countries have these types of systems, this is not the case in developing countries. In developing countries, the metabolites of products used in hospitals or their byproducts, accompanied by a bacterial load (ARGs and HMRGs), are potentially found in hospital effluents treated in situ or collected by urban sewage systems, which are themselves connected to a water treatment plant and discharge the treated effluent into the natural environment (Laffite et al., 2016). In Benin, National Hospital and University Center of Cotonou is the only one hospital with a purification and treatment system for hospital effluents but this system remains moderately efficient (Todedji et al., 2020). Some studies have looked at the microbiological quality of hospital effluents and streams and the isolated bacterial strains have been characterized (Deguenon et al., 2022; Gbotche et al., 2023).
However, none of these studies have evaluated the presence of ARGs and HMRGs in the genome of pathogenic in these different matrices. The present study was undertaken to address this lack of data through. This nationwide study was therefore conducted to determine the current status of ARGs and HMRGs in hospital effluents and waterways in Benin.
MATERIALS AND METHODS
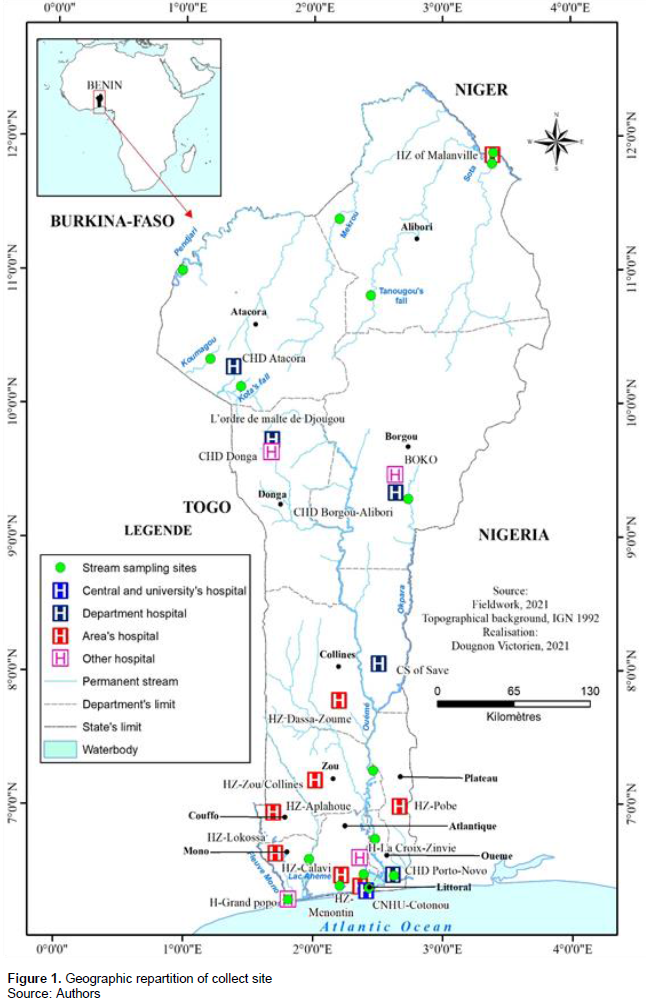
Stream samples were collected from the main streams in Benin. In northern Benin, the Kota and Tanougou waterfalls, the Koumagou, Malanville, Okpara, Sota, Mékrou and Pendjari rivers were sampled. In southern, the rivers of Ganvié, Grand-Popo, Tori, and Porto-Novo, and the lakes of Bopa, Adjohoun, Tokpa, and Zangnannando were sampled (Figure 1). Hospital effluent samples were collected in the National Hospital and University Center (CNHU) of Benin, in the five Departmental Hospital Centers of Benin (CHD Porto-Novo, CHD Borgou-Alibori, CHD Donga and CHD Atacora), in 7 main Zonal Hospitals in the country (HZ Malanville, HZ Dassa-Zoumè, HZ Zou-Collines, HZ Pobè, HZ Aplahoue, HZ Lokossa, HZ Calavi, and HZ Menontin), and in four confessional hospitals in the country (L'ordre de Malte de Djougou, Boko, CS Savè, La croix de Zinvié, and Grand-Popo hospital) (Figure 1).
Two samples were taken from each stream while four samples were collected per site for hospitals effluents. The samples were collected in sterile 1-liter bottles and transported to the laboratoryin a cooler equipped with an accumulator. To target departments with high antibiotic consumption such as intensive care, emergency, pediatrics and maternity departments, collectors were chosen. In total, 32 stream samples and 72 hospital effluent samples were collected from different locations as indicated in Figure 1.
The identification and antimicrobial susceptibility test of multiresistant bacterial strains isolated from these stream and hospitals effluent samples were previously performed and described in the work of Deguenon et al. (2022) and Gbotche et al. (2023).
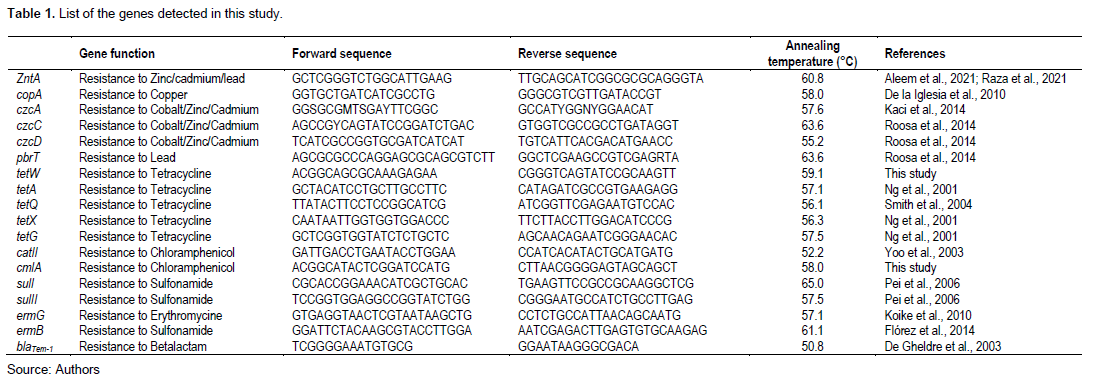
Genomic DNA was extracted from the identified multiresistant drug-resistant bacterial strains using a Quick-DNA TM miniprep kit (Zymo Research Corp, United Stat) according to the manufacturer's instructions. In all, 12 ARGs (cmlA, catII, blaTEM-1, sulI, sulII, tetA, tetQ, tetX, tetG, tetW, ermG, ermB) and six HMRGs (zntA, pbrT, czcA, czcC, czcD, copA) were researched. cmlA, catII, blaTEM-1, sulI, and sulII were researched in gram-negative bacilli, and cmlA, catII, tetA, tetQ, tetX, tetG, tetW, ermG and ermB were researched in gram-positive cocci. Real-time PCR was run using a LineGene9600 Plus Fluorescent Quantitative Detection System (Hangzhou Bioer Technology, China) with the following program: 95°C for 60 s, 40 cycles consisting of (i) 95°C for 15 s, (ii) annealing temperature for 15 s, and a melting stage consisting of (i) 95°C for 15 s, (ii) melting temperature for 60 s and (iii) 95°C for 15 s. Cycle thresholds (CT) were reported, and positive samples were isolated with CT below 30. Primer sequences and annealing temperatures are displayed in (Table 1). Positive controls for antibiotic resistance genes were clinical isolates that carry those genes and the detection was done by standard PCR. For heavy metal genes, to none positive controls were used, but the experiments were ruled twice to confirm the true positive. All negative controls were RNA/DNA free water.
RESULTS
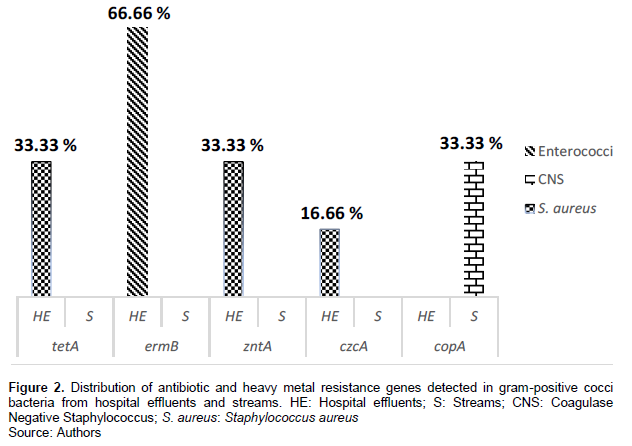
The distribution of ARGs and HMRGs detected in gram-negative bacilli and gram-positive cocci bacteria is as follows. As shown in Figure 2, two ARGs were found in gram-positive cocci bacteria strains, namely, tetA and ermB. tetA was found in 33.33% of the Staphylococcus aureus strains isolated from hospital effluents. While ermB was detected in 66.66% of the Enterococcus strains isolated from hospital effluents. As for the HMRGs detected in gram-positive cocci bacteria, three genes were detected: zntA, czcA, and copA (Figure 2). zntA and czcA were found in 33.33% and 16.66% of the S. aureus strains isolated from hospital effluents, respectively. While, copA was detected in 33.33% of coagulase-negative Staphylococcus (CNS) strains isolated from waterways (Figure 2).
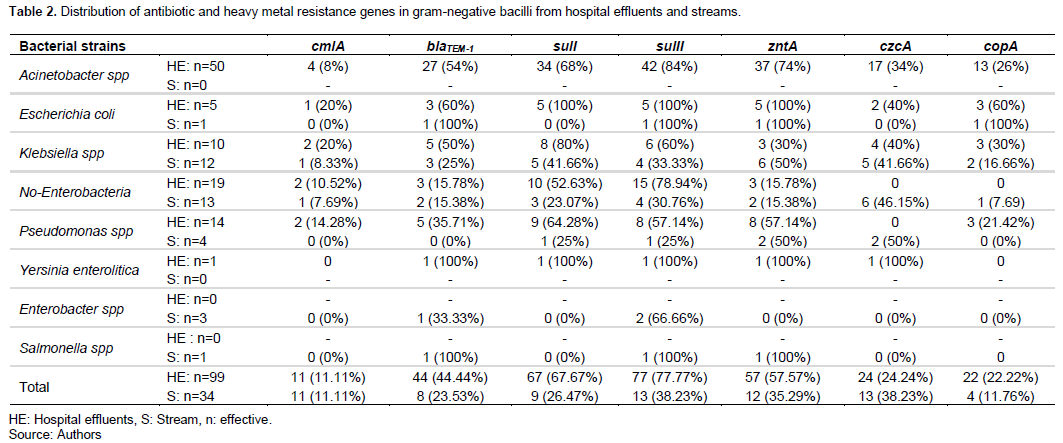
As shown in Table 2, four ARGs (cmlA, blaTEM-1, sulI, and sulII) and three HMRGs (zntA, czcA, and copA) were detected in gram-negative bacilli strains. cmlA was found in 20% of the E. coli and Klebsiella spp. strains; 14.28% of the Pseudomonas spp. strains; and 10.52% of the non-enterobacteria strains, all of which were obtained from hospital effluents. In streams, only 8.33% and 7.69% of Klebsiella spp. and non- enterobacteria strains, respectively, carry the cmlA gene (Table 2). blaTEM-1 was detected in 60% of E. coli strains, 54% of Acinetobacter spp. of Klebsiella spp. strains, and 35.71% of Pseudomonas spp. strains all isolated from hospital effluents. While the same gene was found in 100% of E. coli strains, 33.33% of Enterobacter spp. strains, and 25% of Klebsiella spp. strains, all isolated from streams. The sulI and sulII genes were detected in strains of E. coli, Klebsiella spp. non-enterobacteria and Pseudomonas spp., all isolated both in hospital effluents and in streams (Table 2). Regarding the HMRGs, only the zntA, czcA, and copA genes were detected in the gram-negative bacilli. zntA was found in 100% of E. coli strains, 74% of Acinetobacter spp. strains, and 57.14% of Pseudomonas spp. strains. All isolates were from hospital effluents. The same gene was detected in 100% of E. coli strains, 50% of Klebsiella spp., and Pseudomonas spp. strains, all isolated from streams. czcA was found in the only strain of Yersinia enterolitica, in 40% of strains of E. coli and Klebsiella spp., 34% of strains of Acinetobacter spp. All isolated in the streams. As for the copA gene, it was found in strains of E. coli, Klebsiella spp. isolated both in hospital effluents and in streams (Table 2).
DISCUSSION
The problem of liquid effluent management remains a concern in developing countries like Benin. The objective of this study was to assess the presence of antibiotic and metal resistance gens in Benin hospital effluents and streams. The detection of resistance genes in the extracted DNA of the different bacterial strains isolated showed the presence as well as of ARGs (tetA, blaTEM-1, ermB, sulI, sulII) and HMRGs (zntA, czcA, copA). These genes are found both in hospital effluents (tetA, blaTEM-1, ermB, sulI, sulII, zntA, czcA, copA) and in waterways (cmlA, blaTEM-1, sulI, sulII, zntA, czcA, copA). In hospital effluents, the resistance genes were found in strains of Staphylococcus aureus, coagulase-negative staphylococci, Acinetobacter spp., Escherichia coli, Klebsiella spp., Pseudomonas spp. and Yersinia enterolitica. Bacterial strains of Acinetobacter spp., Yersinia spp., Klebsiella spp., Staphylococcus aureus, and Pseudomonas spp. isolated from wastewater at a sewage treatment plant in western Massachusetts, USA, contained resistance genes to β-lactams, sulfonamides, tetracyclines, zinc, and copper (Martin et al., 2021). Sewage treatment plants receive wastewater from the entire community, including hospital effluents. Therefore, we can say that the results obtained in the present study are in line with those of (Martin et al., 2021). Several studies have shown the presence of antibiotic resistance genes in hospital effluents (Hara et al., 2018; Paul et al., 2018; Yousfi et al., 2019; Fadare and Okoh, 2021), but very few have focused on the presence of HMRGs in effluents. This study provides new scientific data on the presence of heavy metals in hospital effluents. This can be explained by the different human activities practiced and the important flow of humans in hospitals (kitchen, medical care, discharge of heavy metal residues through urine, and feces). These results also support the fact that there is a correlation between the presence of antibiotic and heavy metal resistance genes (Di Cesare et al., 2016). Furthermore, it noted a low presence of blaTEM-1 gene, while it is known that penicillin and cephalosporins are widely used in the country, as indicated by the studies of Dougnon et al. (2020). It would therefore be interesting to update the scientific data on the consumption of antibiotic molecules in Benin. However, it should be noted that the blaTEM-1 genes represent only one of the many genes coding for cephalosporin resistance. It should also be noted that other origins may contribute to the presence of these genes in rivers, including migratory birds in which the same genes have been noted (Yuan et al., 2018). In Poland, and more precisely in the Warmia and Mazury regions, the resistance genes blaTEM-1, sulI, and sulII were detected in river, wastewater and sewage sludge samples (Hubeny et al., 2021). These antibiotic resistance genes were correlated with heavy metals found in variousconcentrations in the same samples (Hubeny et al., 2021). These results are consistent with what have been obtained in this with those in this study, where the presence of blaTEM1, sulI, sulII, and heavy metal resistance genes zntA, czcA, and copA) has been detected. Similar results were obtained by Sabatino et al. (2020) in samples from the Black Sea, where an abundance of tetA, sulII and czcA genes were detected. Hubeny et al. (2021) has reported that wastewater and sewage sludge are discharged into the river. This supports our argument that antibiotic and heavy metal resistance genes are transferred from hospital effluents and community wastewater to Benin's streams. Al Salah et al. (2021) have shown in their studies that the co-occurrence of heavy metals, antibiotic resistant bacteria (ARB), and ARGs in hospital effluent spreading in riverine receiving systems and the assessment of the associated risks are topics of scientific interest and are still little studied in developing countries under tropical conditions.
All these results show the involvement of hospital effluents in the contamination of rivers. It is therefore important that other studies showing the flow of this dissemination should be carried out to identify the treatment and purification sites of hospital effluents before their discharge into streams. This will contribute to the conservation of water resources and help prevent the spread of antimicrobials through a One Health approach.
CONCLUSION
Antibiotic and heavy metal resistance genes are environmental pollutants that contribute significantly to the emergence of multidrug resistance. In the present study, the presence of multidrug-resistant bacteria in hospital effluents was linked to the main streams of Benin. Similar antibiotic resistance genes were found in hospital effluents and streams. These results indicate that hospital effluents are a potential source of dissemination of these hazardous contaminants into water sources. However, it is urgent that these results be used as a basis for monitoring both hospital effluents and streams and for setting up treatment and purification systems for these waters. DNA sequencing to characterize resistance genes and phylogenetic analysis will help to understand and track the flow of antibiotics and metal resistance genes between hospital effluents and streams. However, in the present study, we were not able to carry out these techniques due to the unavailability of the necessary equipment in Benin.
CONFLICT OF INTERESTS
The authors have not declared any conflicts of interests.
ACKNOWLEDGEMENTS
The authors are very grateful to The World Academy of Sciences (TWAS) and the Islamic Development Bank (ISDB), which funded this study. They are finally grateful to the Minister of the Living Environment and Sustainable Development in Benin, his Excellency M. Jose ? TONATO, and all his staff.
FUNDING
The authors are very grateful to The World Academy of Sciences (TWAS) and the Islamic Development Bank (ISDB). These two institutions have made this research possible through research funding allocated to the research team under the IsDB-TWAS Grants for Research Collaboration in Sustainability Sciences (506808). They reviewed the research protocol and validated the design of the study and collection, analysis, and interpretation of data.
REFERENCES
|
Adelowo OO, Caucci S, Banjo OA, Nnanna OC, Awotipe EO, Peters FB, Fagade OE, Berendonk TU (2018). Extended Spectrum Beta-Lactamase (ESBL)-producing bacteria isolated from hospital wastewaters, rivers and aquaculture sources in Nigeria. Environmental Science and Pollution Research 25:2744-2755. |
|
|
Al Salah DMM, Laffite A, Sivalingam P, Poté J (2021). Occurrence of toxic metals and their selective pressure for antibiotic-resistant clinically relevant bacteria and antibiotic-resistant genes in river receiving systems under tropical conditions. Environmental Science and Pollution Research 29(14):20530-20541. |
|
|
Bartley PS, Domitrovic TN, Moretto VT, Santos CS, Ponce-Terashima R, Reis MG, Barbosa LM, Blanton RE, Bonomo RA, Perez F (2019). Antibiotic resistance in Enterobacteriaceae from surface waters in urban Brazil highlights the risks of poor sanitation. The American Journal of Tropical Medicine and Hygiene 100(6):1369-1377. |
|
|
Carles L, Gardon H, Joseph L, Sanchís J, Farre M, Artigas J (2019). Meta-analysis of glyphosate contamination in surface waters and dissipation by biofilms. Environment international 124:284-293. |
|
|
Chen J, Li J, Zhang H, Shi W, Liu Y (2019). Bacterial heavy-metal and antibiotic resistance genes in a copper tailing dam area in northern China. Frontiers in Microbiology 10:1916. |
|
|
Dahanayake PS, Hossain S, Wickramanayake M, Heo GJ (2019). Antibiotic and heavy metal resistance genes in Aeromonas spp. isolated from marketed Manila Clam (Ruditapes philippinarum) in Korea. Journal of Applied Microbiology 127(3):941-952. |
|
|
De Gheldre Y, Avesani V, Berhin C, Delmée M, Glupczynski Y (2003). Evaluation of Oxoid combination discs for detection of extended-spectrum β-lactamases. Journal of Antimicrobial Chemotherapy 52(4):591-597. |
|
|
Deguenon E, Dougnon V, Houssou VMC, Gbotche E, Ahoyo RA, Fabiyi K, Boko M (2022). Hospital effluents as sources of antibiotics residues, resistant bacteria and heavy metals in Benin. SN Applied Sciences 4(8):206. |
|
|
De la Iglesia R, Valenzuela-Heredia D, Pavissich JP, Freyhoffer S, Andrade S, Correa JA, González B (2010). Novel polymerase chain reaction primers for the specific detection of bacterial copper P-type ATPases gene sequences in environmental isolates and metagenomic DNA. Letters in Applied Microbiology 50(6):552-562. |
|
|
Di Cesare A, Eckert EM, D'Urso S, Bertoni R, Gillan DC, Wattiez R, Corno G (2016). Co-occurrence of integrase 1, antibiotic and heavy metal resistance genes in municipal wastewater treatment plants. Water Research 94:208-214. |
|
|
Dougnon V, Chabi Y, Koudokpon H, Agbankpe J, Sefounon R, Alle D, Bankolé H, Baba-Moussa L (2020). Prescription of antibiotics as a source of emerging antibiotic resistance: Knowledge, attitudes, and practices of medical staff in the Dassa-Glazoué and Savalou-Bantè's health zones (Benin, West Africa). International Journal of One Health 6:2455-8931. |
|
|
Duan K, Zhao B, Zhang S, Ma Y (2021). Contamination characteristics, source analysis, and ecological risk assessment of toxic metals and metalloid in agricultural soil in Yuzhong, China. Wiley Online Library. |
|
|
Fadare FT, Okoh Al (2021). Distribution and molecular characterization of ESBL, pAmpC β-lactamases, and non-β-lactam encoding genes in Enterobacteriaceae isolated from hospital wastewater in Eastern Cape Province, South Africa. Plos One 16(7):e0254753. |
|
|
Flórez AB, Alegría Á, Rossi F, Delgado S, Felis GE, Torriani S, Mayo B (2014). Molecular identification and quantification of tetracycline and erythromycin resistance genes in Spanish and Italian retail cheeses. BioMed research international. |
|
|
Gbotche E, Houssou Quenum MC, Dougnon TV, Ogunlaja A, Klotoe JR, Fabiyi K, Unuabonah IE (2023). National Survey of Stream Water Quality Revealing Threats to Antibio-Resistant Bacteria, Antibiotic Residues and Heavy Metals in Benin. Pollution 9(2):678-692. |
|
|
Giannakis S, Rtimi S, Pulgarin C (2017). Light-assisted advanced oxidation processes for the elimination of chemical and microbiological pollution of wastewaters in developed and developing countries. Molecules 22(07):1070. |
|
|
Gogry FA, Siddiqui MT (2019). Emergence of mcr-1 conferred colistin resistance among bacterial isolates from urban sewage water in India. Environmental Science and Pollution Research 26(32):33715-33717. |
|
|
Gogry FA, Siddiqui MT, Sultan I, Haq QMR (2021). Current update on intrinsic and acquired colistin resistance mechanisms in bacteria. Frontiers in Medicine 8:677-720. |
|
|
Gogry FA, Siddiqui MT, Sultan I, Husain FM, Al-Kheraif AA, Ali A, Haq QMR (2022). Colistin interaction and surface changes associated with mcr-1 conferred plasmid mediated resistance in E. coli and A. veronii strains. Pharmaceutics 14(2):295. |
|
|
Hara H, Yusaimi YA, Zulkeflle SNM, Sugiura N, Iwamoto K, Goto M, Utsumi M, Othman N bin, Zakaria Z (2018). Molecular characterization of multi-drug resistant Escherichia coli isolates from tropical environments in Southeast Asia. The Journal of General and Applied Microbiology Advpub 64(6):284-292. |
|
|
Hubeny J, Harnisz M, Korzeniewska E, Buta M, Zieli?ski W, Rolbiecki D, Giebultowicz J, Nal?cz-Jawecki G, Plaza G (2021). Industrialization as a source of heavy metals and antibiotics which can enhance the antibiotic resistance in wastewater, sewage sludge and river water. PloS One 16:e0252691. |
|
|
Kaci A, Petit F, Lesueur P, Boust D, Vrel A, Berthe T (2014). Distinct diversity of the czcA gene in two sedimentary horizons from a contaminated estuarine core. Environmental Science and Pollution Research 21:10787-10802. |
|
|
Kayembe JM, Sivalingam P, Salgado CD, Maliani J, Ngelinkoto P, Otamonga J-P, Mulaji CK, Mubedi JI, Poté J (2018). Assessment of water quality and time accumulation of heavy metals in the sediments of tropical urban rivers: Case of Bumbu River and Kokolo Canal, Kinshasa City, Democratic Republic of the Congo. Journal of African Earth Sciences 147:536-543. |
|
|
Koike S, Aminov RI, Yannarell AC, Gans HD, Krapac IG, Chee-Sanford JC, Mackie RI (2010). Molecular ecology of macrolide-lincosamide-streptogramin B methylases in waste lagoons and subsurface waters associated with swine production. Microbial Ecology 59:487-498. |
|
|
Laffite A, Kilunga PI, Kayembe JM, Devarajan N, Mulaji CK, Giuliani G, Slaveykova VI, Poté J (2016). Hospital Effluents Are One of Several Sources of Metal, Antibiotic Resistance Genes, and Bacterial Markers Disseminated in Sub-Saharan Urban Rivers. Frontiers in Microbiology 7:1128. |
|
|
Laffite A, Al Salah DMM, Slaveykova VI, Otamonga J-P, Poté J (2020). Impact of anthropogenic activities on the occurrence and distribution of toxic metals, extending-spectra β-lactamases and carbapenem resistance in sub-Saharan African urban rivers. Science of the Total Environment 727:138129. https://doi.org/10.1016/j.scitotenv.2020.138129 |
|
|
Mandaric L, Kalogianni E, Skoulikidis N, Petrovic M, Sabater S (2019). Contamination patterns and attenuation of pharmaceuticals in a temporary Mediterranean river. Science of the Total Environment 647:561-569. |
|
|
Martin C, Stebbins B, Ajmani A, Comendul A, Hamner S, Hasan NA, Colwell R, Ford T (2021). Nanopore-based metagenomics analysis reveals prevalence of mobile antibiotic and heavy metal resistome in wastewater. Ecotoxicology 30:1572-1585. |
|
|
Mittelman MW, Jones AD (2018). A pure life: the microbial ecology of high purity industrial waters. Microbial Ecology 76(1):9-18. |
|
|
Ng LK, Martin I, Alfa M, Mulvey M (2001). Multiplex PCR for the detection of tetracycline resistant genes. Molecular and Cellular Probes 15:209-215. |
|
|
Niest?pski S, Harnisz M, Korzeniewska E, Osi?ska A (2020). Markers Specific to Bacteroides fragilis Group Bacteria as Indicators of Anthropogenic Pollution of Surface Waters. International Journal of Environmental Research and Public Health 17(19):7137. |
|
|
Ohore OE, Addo FG, Zhang S, Han N, Anim-Larbi K (2019). Distribution and relationship between antimicrobial resistance genes and heavy metals in surface sediments of Taihu Lake, China. Journal of Environmental Sciences 77:323-335. |
|
|
Paul C, Bayrychenko Z, Junier T, Filippidou S, Beck K, Bueche M, Greub G, Bürgmann H, Junier P (2018). Dissemination of antibiotic resistance genes associated with the sporobiota in sediments impacted by wastewater. Peer J 6:e4989. |
|
|
Pei R, Kim S-C, Carlson KH, Pruden A (2006). Effect of river landscape on the sediment concentrations of antibiotics and corresponding antibiotic resistance genes (ARG). Water Research 40(12):2427-2435. |
|
|
Pietryczuk A, Cudowski A, Hauschild T, ?wislocka M, Wi?cko A, Karpowicz M (2018). Abundance and species diversity of fungi in rivers with various contaminations. Current Microbiology 75:630-638. |
|
|
Posada-Perlaza CE, Ramírez-Rojas A, Porras P, Adu-Oppong B, Botero-Coy A-M, Hernández F, Anzola J.M, Díaz L, Dantas G, Reyes A (2019). Bogotá River anthropogenic contamination alters microbial communities and promotes spread of antibiotic resistance genes. Scientific Reports 9:1-13. |
|
|
Raza S, Shin H, Hur HG, Unno T (2021). Higher abundance of core antimicrobial resistant genes in effluent from wastewater treatment plants. Water Research 208:117882. |
|
|
Roosa S, Wattiez R, Prygiel E, Lesven L, Billon G, Gillan DC (2014). Bacterial metal resistance genes and metal bioavailability in contaminated sediments. Environmental Pollution 189:143-151. |
|
|
Sabatino R, Di Cesare A, Dzhembekova N, Fontaneto D, Eckert EM, Corno G, Moncheva S, Bertoni R, Callieri C (2020). Spatial distribution of antibiotic and heavy metal resistance genes in the Black Sea. Marine Pollution Bulletin 160:111635. |
|
|
Sheu CC, Chang YT, Lin SY, Chen YH, Hsueh PR (2019). Infections caused by carbapenem-resistant Enterobacteriaceae: an update on therapeutic options. Frontiers in Microbiology 10:80. |
|
|
Smith MS, Yang RK, Knapp CW, Niu Y, Peak N, Hanfelt MM, Galland JC, Graham DW (2004). Quantification of tetracycline resistance genes in feedlot lagoons by real-time PCR. Applied and Environmental Microbiology 70:7372-7377. |
|
|
Suzuki Y, Nazareno PJ, Nakano R, Mondoy M, Nakano A, Bugayong MP, Bilar J, Perez M, Medina EJ, Saito-Obata M (2020). Environmental presence and genetic characteristics of carbapenemase-producing Enterobacteriaceae from hospital sewage and river water in the Philippines. Applied and Environmental Microbiology 86: e01906-19. |
|
|
Todedji JN, Degbey CC, Soclo E, Yessoufou A, Goudjo F, Hounfodji JW, Suanon F, Mama D (2020). Caractérisation physico-chimique et toxicologique des effluents des Centres Hospitaliers et Universitaires du département du Littoral du Bénin. International Journal of Biological and Chemical Sciences 14(3):1118-1132. |
|
|
Turner RJ, Huang L-N, Viti C, Mengoni A (2020). Metal-Resistance in Bacteria: Why Care? Genes 11(12):1470. |
|
|
Verburg I, García-Cobos S, Hernández Leal L, Waar K, Friedrich AW, Schmitt H (2019). Abundance and antimicrobial resistance of three bacterial species along a complete wastewater pathway. Microorganisms 7(9):312. |
|
|
Yeh G, Hoang HG, Lin C, Bui XT, Tran HT, Shern CC, Vu CT (2020). Assessment of heavy metal contamination and adverse biological effects of an industrially affected river. Environmental Science and Pollution Research 27(28):34770-34780. |
|
|
Yoo MH, Huh MD, Kim E, Lee HH, Jeong HD (2003). Characterization of chloramphenicol acetyltransferase gene by multiplex polymerase chain reaction in multidrug-resistant strains isolated from aquatic environments. Aquaculture 217:11-21. |
|
|
Yousfi K, Touati A, Lefebvre B, Garneau P, Brahmi S, Gharout-Sait A, Harel J, Bekal S (2019). Characterization of multidrug-resistant Gram-negative bacilli isolated from hospitals effluents: first report of a blaOXA-48-like in Klebsiella oxytoca, Algeria. Brazilian Journal of Microbiology 50:175-183. |
|
|
Yuan QB, Zhai YF, Mao BY, Hu N (2018). Antibiotic resistance genes and intI1 prevalence in a swine wastewater treatment plant and correlation with metal resistance, bacterial community and wastewater parameters. Ecotoxicology and Environmental Safety 161:251-259. |
|
|
Zhou XY, Wang X (2019). Cd contamination status and cost-benefits analysis in agriculture soils of Yangtze River basin. Environmental Pollution 254:112962. |
|
Copyright © 2024 Author(s) retain the copyright of this article.
This article is published under the terms of the Creative Commons Attribution License 4.0