ABSTRACT
The incidence of meningoencephalitis in developing countries is high and its diagnosis being inadequate as it is mainly based on conventional methods with a limited diagnostic capacity for bacterial, fungal and viral detection. This results in prescription of a cocktail of antibiotics, antifungal, or anti-viral drugs, since the causative agent is often unknown. There is therefore need for timely and appropriate diagnosis to guide treatment. This study aims at evaluating the utility of molecular diagnostic over conventional methods in detection of etiologic agents of central nervous system infections in Rwanda. Using a cross sectional design, 845 hospitalized patients suspected to have meningoencephalitis were enrolled from 8 study sites across the country. Four sterile tubes of Cerebral Spinal Fluid (CSF) specimens were collected from each patient. Two tubes were analyzed on site for bacteriology, fungal, biochemistry and cytology while the other two were transported to the National Reference Laboratory (NRL) for culture and Real-Time Multiplex Polymerase Chain Reaction (PCR) assays. Data entry and analysis was done using Epi Info7, Excel; SPSS and STATA16. In the study, no viruses were detected using the conventional methods while a range of viruses, 152/845 (18%) were detected using real time multiplex PCR. In addition, 185/845 (22%) samples were detected as positive for different types of bacteria using the PCR compared with the 59/845(7%) that were positive using conventional methods. There was however no difference in the detection capacity of fungal agents between the two methods with a detection level of 6% (49). This study has shown increased capacity for detection of bacterial and viral causative agents of Central Nervous System (CNS) infections using RT multiplex PCR compared with conventional methods. This result facilitated the development of a novel algorithm for both conventional and molecular diagnostic methods for CNS infections.
Key words: Meningoencephalitis, RT- Multiplex PCR, conventional methods, causative agents, diagnostic capacity, cerebral spinal fluid.
The etiology of CNS infections varies by age and place. According to previous studies, bacterial meningitidis accounts for approximately 30 to 40% of CNS infections while the remaining 60 to 70% are caused by cryptococcal, viral, protozoa, brain abscess and other etiologic agents (Mengistu et al., 2013).
In Sub-saharan Africa, Neisseria meningitidis infection rates can be as high as 1 in 1000 to 1 in 100 as compared to about 1 in 100,000 in United States (Richardson et al., 2002). More than 90% of Neisseria meningitidis infections in Sub-Saharan Africa are caused by serogroups Y, W135, C, B, and A.
Meningoencephalitis is a serious disease resulting from an acute inflammation of
meninges which causes significant morbidity and mortality (Baskin and Hedlund, 2007).
At the initial stage of meningoencephalitis, no reliable clinical indicators are available to differentiate between bacterial, viral, fungal or protozoal infections. As a result, all suspected cases of meningoencephalitis are normally referred for hospitalization. It is very important to identify the causative agents of the meningoencephalitis at an early stage, and determine the appropriate treatment in order to minimize unnecessary antibiotic, antiviral, antifungal and antiparasitic prescriptions (Baskin and Hedlund, 2007).
Bacterial meningitides is mostly caused by Streptococcus pneumoniae, Haemophilus influenza, Neisseria meningitidis, Listeria monocytogenes and group B Streptococci (Bartt, 2012). In children, the common causes of meningococcal infection are Haemophilus influenzae type b (Hib) and S. pneumoniae (Taylor et al., 2012).
Viral meningitidis is mostly caused by Enteroviruses and is associated with neurological complications and mortality in 35 to 83%, followed by mumps distant second with 1 to 40% (Minjolle et al., 2002). Other viruses that also cause CNS disease are herpes viruses, toga viruses, bunya viruses, lymphocytic choriomeningitis virus, measles and rubella viruses.
Cytomegalovirus (CMV) infections occur mostly in patients with AIDS causing sub-acute encephalitis. Congenital CMV, which is a much more serious form of infection, has a significant association with morbidity and mortality (Bartt, 2012). In immunocompetent individuals, adenovirus is a rare cause of meningitidis but it remains a key cause of upper respiratory infections in immunocompromised persons (Cho and Mckendall, 2014). Early treatment with acyclovir can significantly reduce morbidity caused by CMV (Bartt, 2012).
In the past, diagnosis of viral meningoencephalitis has been difficult, but it is improving with newer molecular methods (Logan and MacMahon, 2008).The clinical outcome of viral encephalitis is favorable when treatment is done early. Therefore, underlining the importance of early specific diagnosis of meningitis and encephalitis (Baskin and Hedlund, 2007).
Fungi cause severe infections but are much less frequent than bacterial or viral infections. The most common causes of fungal meningoencephalitis are Cryptococcus neoformans, Candida albicans and Aspergillus species especially in immunocompromised patients (Baskin and Hedlund, 2007).
Conventional diagnostic methods relying on isolation of bacteria from CSF has previously been shown to be unreliable due to time of culture and prior administration of antibiotics before CSF collection (Ntagwabira et al., 2017). Taken together, there is a need for prompt molecular diagnosis of meningoencephalitis so as to guide early and appropriate treatment and avoid clinical scenarios whereby a patient receives empirically, a cocktail of antibiotics in the absence of causative agent detection.
The sample collection was conducted between February 2017 and February 2018 on 845 consecutively sampled inpatients from four referrals, and four satellite hospitals with bacteriology laboratories namely King Faisal, Rwanda Military, Butare University Teaching and Kigali University Teaching Referral Hospitals; and four satellites hospitals namely, Gisenyi, Kibungo, Nyagatare and Byumba satellite hospitals.
The study teams composing of medical doctors, nurses and laboratory personnel were recruited from all the sites and centrally trained at the National Reference laboratory on the criteria for patient recruitment, sample collection, storage and transportation.
All consenting and assenting inpatients and coma patients’ proxies of all ages with clinical signs and symptoms of meningoencephalitis were recruited into the study.
Patients with International Normalized Ratio (INR) of less than 1.5 and those with a platelet count of less than 50,000 were excluded. Similarly, those with intracranial mass and partial/complete spinal block or acute spinal trauma were also excluded.
The physicians in the study sites were responsible for initial clinical diagnosis and selection of meningoencephalitis patients to be enrolled in the study based on case definition and the most frequent common symptoms were not limited to fever, headache, cranial nerve abnormalities and chills, confusion, irritability and neck stiffness (Table 2). Each patient’s details were entered into case report form detailing their medical history and the socio-demographic information.
CSF was collected into four individual sterile screw cup tubes with a minimum of 1 ml per tube using standard CSF collection techniques and labelled with the study unique identification number.
The CSF from one tube was used for gram stain, Indian ink stain for cryptococcal antigen (Crag) detection and conventional culture at site. Another tube was used for the biochemistry and cytology analysis within one hour. CSF sample of 0.5-1 ml from one of the remaining two tubes was inoculated into Trans-Isolate (TI) culture medium while the other one was stored at - 20°C waiting for transportation to NRL for pathogen detection using culture and RT-multiplex PCR, respectively. During inoculation of the CSF to the media, contamination was avoided by using sterile forceps to remove the aluminum foil covering the medium and disinfecting the stopper with 70% alcohol and allowed to dry. Sterile syringes and needles were used during the inoculation.
If transportation for TI culture medium to NRL was delayed to the next day or longer, averting needle (sterile cotton plugged hypodermic needle) was inserted through the rubber stopper of the T-I medium bottle, which ensured growth and survival of the bacteria.
CSF specimens were handled as potentially infectious. Biosafety guidelines were adhered by using biosafety cabinets during manipulation of the specimens according to the World Health Organization (WHO) and National Reference Laboratory Guidelines.
Sample processing at NRL using conventional method
Each sample has been transported with its consent or assent form and case report form within 4 h after sample collection.
Upon arrival at NRL, the sample in T-I bottle was aseptically inoculated on sheep blood agar, chocolate agar and MacConkey agar. The remaining sample was incubated at 35-37°C in a ~5% CO2 atmosphere then observed daily for turbidity in the liquid phase for up to 7 days to enhance the growth if the first subculture did not show any growth. The venting needle was inserted in the T-I bottle (sterile cotton-plugged hypodermic needle) through the rubber stopper of the T-I bottle without touching the broth, in order to encourage growth and survival of bacteria.
When T-I became turbid, the sample was cultured on blood agar and chocolate agar incubated at 35-37°C in a ~5% CO2 atmosphere for 24 h, culture was also done on MacConkey agar and 35-37°C for 18 to 24 h. The T-I at this time was discarded. If no turbidity was observed after 7 days, sample was systematic inoculated on blood agar, chocolate agar and Mac Conkey agar, at this point the T-I was discarded and the final culture was recorded and reported to attending physician.
On sheep blood agar and chocolate agar if there was no growth observed in 24 h, the plates were re-incubated for 48 h before reporting negative culture. The culture growth, identification of colonies was done using Gram staining of young colonies, followed by biochemical testing (API 20E, API NH, catalase, coagulase, oxidase, CAMP test) and serotyping. After bacterial identification antimicrobial susceptibility testing was conducted to all isolated bacteria using recommended antibiotic discs on Mueller-Hinton agar, Kirby-Bauer method and Clinical and Laboratory Standards Institute (CLSI) catalogue for result interpretation.
A small quantity of CSF sample for RT Multiplex PCR was aseptically inoculated on Yeast Extract Peptone Dextrose (YPD) medium and incubated at 35- 37°C; the growth was checked after 48 h. If no growth, the plate was re-incubated and checked daily for up to 7 days. The remaining sample was tightly closed and stored at -20°C for RT multiplex PCR. The colonies growing on YPD were identified using Indian ink and Cryptococci antigen latex agglutination.
The results were recorded on a worksheet, then case report form and entered into Laboratory Information System and Epi-Info database. All positive results were communicated to attending physician immediately by phone call and all printed results were sent by sample transportation car going to each hospital once weekly or given to research site laboratory technologist under research team bringing samples at NRL.
Quality assurance on conventional method
Quality assurance system and continuous monitoring was built to ensure that all procedures were performed proficiently, adequately and correctly.
Quality, accuracy and reliability of the study results on conventional method was validated and verified based on the NRL-External Quality Control (EQC) policy. The known positive samples were selected from the NRL archived strains. The available strains for positive control were N. meningitidis, S. pneumoniae, and H. influenzae B, from the NHLS, South Africa and C. neoformans from NRL archive as well as ATCC bacterial species (S. pneumoniae, E. coli and H. influenzae B). All samples were processed according to the Standard Operating Procedures.
Samples analysis using RT Multiplex PCR
RT multiplex PCR assay validation
Quality control
Known positive control strains used during culturing above were also used as the positive control for the bacteria detection and specificity of the PCR assays. The strains had been sub-cultured from the skim milk and thus DNA extraction had to be performed.
The reliability of the study was validated and verified based on the NRL-EQC policy. The known positive samples were selected from the NRL archived strains. The available strains for positive control were N. meningitidis, S. pneumoniae, and H. influenzae B, from the NHLS, South Africa and C. neoformans from NRL archive as well as ATCC bacterial species (N. meningitis, S. pneumoniae, E. coli and H. influenzae B).
Nucleic acid extraction
Nucleic acid was extracted from the known positive controls: N. meningitidis, S. pneumoniae, H. influenzae B, C. neoformans and E. coli) using an automated nucleic acid extraction platform (QIAsymphony SP-QIAGEN, Germany). The QIAsymphony DSP virus /pathogen mini kit was used according to manufacturer’s instruction with a starting volume of 250 μl and an elution volume of 60 μl. The QIAsympony SP uses the magnetic bead technology, where the viral/pathogen cells are first lysed to release the nuclei acid. The released nucleic acid binds to the magnetic particles allowing separation of the nucleic acid.
The nucleic acid was then purified by washing in the series of wash buffer on rod covers before finally being released in the elution buffer. Sterile nuclease free water was used as a negative control of extraction.
The extracted nucleic acid was collected in elution racks, labelled and quantified using QiAxpert nucleic acid quantifier system (QIAGEN, Germany) to establish the quality and quantity of the eluates. The quality of extracted nucleic acid was validated on wavelengths of 230/260 and 260/280 ratios to check for sample purity. The eluted nucleic acid was used directly in Real time-PCR and the excess stored at -80°C for long storage or 4 to 8°C for short storage.
Master mix preparation
Pre-optimized Commercial RT-Multiplex PCR kits for in-vitro diagnostics were used. The kits comprising PCR buffer, primer and probe mix (PPmix) and Taq polymerase enzyme were provided in three separate tubes and required constitution of the master mix before adding the templates. Both automated and manual master mix preparations were evaluated for their efficiency in processes and also on optimal reagent usage. QIAgility (QIAGEN, Germany) liquid handler was used for the master mix. The system was programmed to pipette 12.5 µl of PCR buffer, 1.5 µl of PPmix, 1.5 µl of the enzyme and 2 µl of internal control into a 1.5 ml tube and later aliquoted 15 µl into individual 0.1 ml PCR ml tubes that were compatible with the thermalcycler (Rotor Gene Q, QIAGEN- Germany). 10 ml of the extracted sample nucleic acid was added to the master mix into each tube including the extracted negative control sample. Positive and negative controls from the manufacturer were included in each run. In total, four RT-Multiplex PCR kits were used namely: FTD bacterial meningitis (N. Meningitidis, S. pneumoniae and H. influenzae)-Fast Track Diagnostics, Luxembourg, FTD Neonatal meningitis kit (S. agalactiae/Group B Streptococcus, Listeria monocytogenes and E. coli)- )-Fast Track Diagnostics, Luxembourg, C. neoformans Real-TM kit (C. neoformans)-Sacace Biotechnologies, Italy and FTD Neuro9 kit (human Adenovirus-ADV, human Cytomegalovirus-CMV, Epstein-Barr virus-EBV, Herpes simplex virus-HSV, 1 and 2, Varicella zoster virus -VZV, Enterovirus, human parechovirus -HPV, human herpesvirus 6 and 7, and human parvovirus B19)- )-Fast Track Diagnostics, Luxembourg. The PCR setup for the Cryptococcus neoformans entailed 10 µl of PCR mix -1, 1.5 µl of PCR mix-2, 0.5 µl of DNA Polymerase, 2 µl of internal control and sterile water into each PCR reaction tube followed by 10 µl of the extracted nucleic acid. Specificity of the assays was tested by amplifying known strains in the kits without their targets. The tubes were sealed, labelled and transferred to the real time thermalcycler (Rotor Gene Q -, Germany) for amplification.
Amplification
The commercial pre-optimized RT Multiplex PCR kits contain all the necessary reagents for amplification. Each primer and probe mix has been specifically designed to target specific pathogen targets and validated for specificity but checking the possibility to amplify related pathogens. In the case of viruses, the viral RNA is transcribed into cDNA using a specific primer mediated reverse transcription step followed immediately in the same tube by polymerase chain reaction. The DNA is amplified simultaneously in the same tube by polymerase chain reaction. In situations of multiplex PCR, the reaction will have a cocktail of primers and probes targeting multiple pathogens. The presence of specific pathogen sequences in the reaction is detected by an increase in fluorescence observed from the relevant dual-labeled probe, and reported as a cycle threshold value (Ct) by the Real-time thermocycler. The assays use murine Cytomegalovirus (mCMV) and Brome mosaic virus (BMV) as the internal control. The internal control is introduced into each PCR reaction tube during the PCR setup to check for PCR efficiency.
The kits use similar amplification programme involving an initial enzyme activation step at 95°C for 15 min and 5 cycles comprising of denaturation at 95°C for 5 s, annealing step at 60°C for 20 s and extension step at 72°C for 15 s without fluorescence detection.
This was followed by 40 cycles comprising denaturation at 95°C for 5 s, annealing step at 600C for 15 s and extension step at 72°C for 15 s with fluorescence detection on four detection channels of the thermocycler as indicated in Table 2.
Similarly, the pre-optimized commercial Real-Time PCR kits was used for detection of Cryptococcus neoformans and entailed an initial enzyme activation step at 95°C for 15’ min and 5 cycles comprising denaturation at 95°C for 5 seconds, annealing step at 60°C for 20 s and extension step at 72°C for 15 s without fluorescence detection, followed by 40 cycles comprising denaturation at 95°C for 5 s, annealing step at 60°C for 20 s and extension step at 72°C for 15 s with fluorescence detection on green and yellow at the annealing step at 60°C
Interpretation of the PCR assays
Specific probes were detected on four different channels on Rotor Gene Q which were matched up with specific pathogens as shown in Table 1. The detection involves analyzing the PCR assay upon completion of the run to check the validity of the assay by confirming absence of any contamination and establishing PCR amplification efficiency. The process entails using the Rotor Gene Q software to first check for contamination by analyzing the negative controls. The threshold is set at a point where the negative control is below the threshold and the Ct value for the positive and internal controls is below 33 while the amplification exhibits positive (exponential) trace as guided by the manufacturer instructions commercial kits. Any specimen displaying an exponential trace with a Ct value below 33 is considered positive. A detection guide outlined in Table 1 was provided by the manufacturer for establishing the pathogens detected in the positive samples. Appearance of any curve in the negative control was considered as potential contamination, and therefore the results obtained were not interpretable. The whole run including extraction had to be repeated. The run was also repeated where the positive and internal controls did show amplification curves.
The validation of the RT multiplex PCR Assays was considered successful upon detection of the known NRL strains used as positive controls of nucleic acid purification. The assays also amplified the positive and internal controls provided in the kits while both the negative control of purification and the one provided in the kit did not amplify.
Table 1 shows the detection guide for the targets in the four RT PCR kits used for analysis of 845 samples from patient suspect to have CNS infections.
Upon validation of the assays, nucleic acid from the patients’ samples were extracted, quantified and amplified as per the above validated protocols with reference to manufacturer’s instructions.
Ethical consideration
This study was reviewed and approved by both Rwanda National Ethical Committee and Kenyatta National Hospital/University of Nairobi Ethics and Research Committee prior to its commencement. The approval numbers are No 572/RNEC/2041, KNH/UON ERC-P561/09/2014 respectively. Written informed consent was obtained from all patients or proxies of comatose patients aged 18 years and above. In addition, assent was obtained from parents/guardians of children aged below 18 years by the signing of assent forms. The study was explained to the study participants by the nurses at the sites to ensure that they understood the significance, design and objectives and any risks that may be involved. Privacy and confidentiality of the patients was upheld at all stages of the study. All study subjects were informed that they were free to withdraw their participation from the study at any stage without any loss of benefits or penalty. Personal identifiers were removed from all the data and replaced with unique numbers.
A total of 845 CSF samples obtained from patients suspected to be having meningoencephalitis were analyzed in this study using conventional and molecular diagnostic methods. The total number of female patients was 434 (51.4%) of the study population while male patients were 411(48.6%) of the study population.
Characteristics of study population
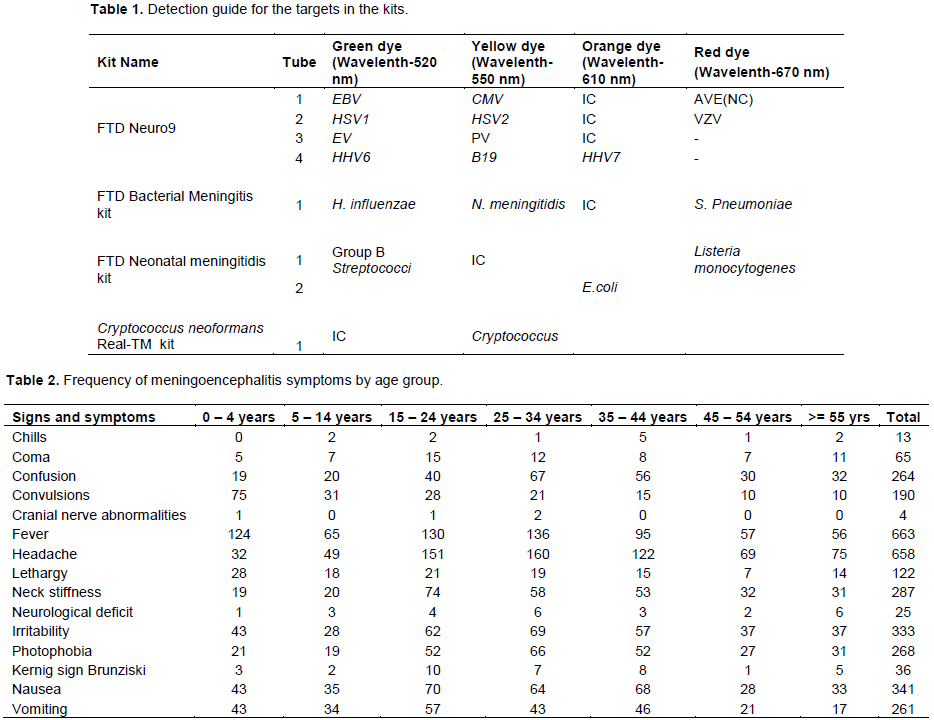
Figure 1 shows the demographic representation of the age groups in the study. About 65% of the study population was below 35 years of age. The most frequently affected population was the age group of 25 to 34 years’ old which constituted 21% of the patients seen followed by group aged 15 to 20 years which constituted 20% of the patients. The least frequently affected age group was that aged 5 to 14 years old constituting 9% of the patient population.
Clinical characteristics of study population
Table 2 shows the frequency of signs and symptoms in patients suspected to have meningoencephalitis. The most frequently observed sign was fever, seen in 663 (75.8%) of the patients while headache was the most frequently reported symptom by 658 (75.2%) of the patients. Only 4(0.5%) and 13(1.5%) had cranial nerve abnormalities or chills, respectively.
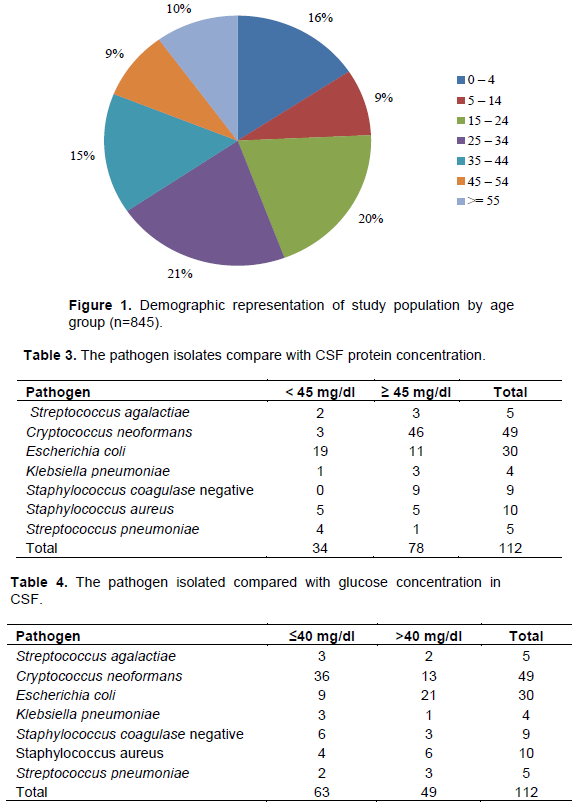
Table 3 indicates that according to protein concentration in CSF samples, there are two groups: one with protein below 45 mg/dl and another one with protein equal to 45 mg/dl and above. The normal protein concentration in CSF is below 45 mg/dl.
Isolated Staphylococcus coagulase negative 9/ 9 (100%) showed significantly higher CSF protein concentration than other isolated microorganisms, followed by C. neoformans 46 / 49 having significantly higher CSF protein concentration (93.9%).
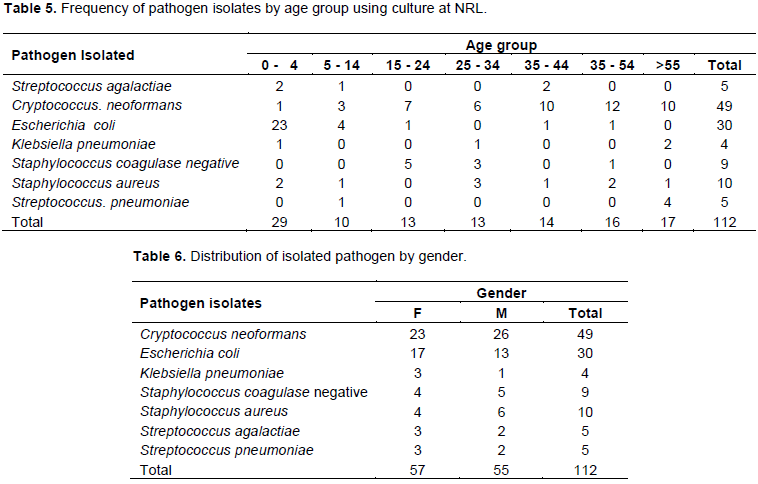
Table 4 shows the normal glucose concentration in CSF is between 45–80 mg/dl. The patients with bacterial
Outcome of culture at NRL
The results from culture at NRL showed that out of 845 cultured samples, only 63(7%) samples were positive to different types of bacteria while 49(6%) were positive to C. neoformans as indicated in Table 5. In this study there was no virus culture facility, therefore no viruses were detected using conventional method.
Table 5 shows positivity rates for the 845 CSF samples cultured at NRL, whereby 63/8457% were positive with bacteria. The most isolated bacteria were E. coli 30/63 (48%), which was more isolated from children between 0 to 4 years, followed by S. aureus 10 (16%), S. coagulase negative 9 (14%), S. agalactiae 5(8%), S. pneumoniae 5 (8%); and K. pneumoniae, 4(6%). Similarly, 49 samples out of 845 (6%) were positive for C. neoformans.
Table 6 shows the pathogens isolated from culture compared in relationship to gender. There is no significant difference in the isolated pathogens from men and women, since isolates from men were 55/411 (13%) and that from women were 57/434 (13%).
Outcome of RT-Multiplex PCR at NRL
Using the developed and validated RT-Multiplex PCR algorithm, 845 CSF samples were collected and analyzed. 152 (18%) were positive for the detected viruses below: Adenovirus 38/ 845 (5%), Epstein-Barr virus 22 / 845 (3%), Cytomegalovirus 7/845 (1%), Human Herpes virus (6) 17/845 (2%), Human Herpes virus (7) 3/ 845(0%), Herpes simplex virus (1) 6 /845 (1%) and Herpes simplex virus (2) 3 / 845 (0%), Enterovirus 52 /845 (6%) and Human Parechovirus 4 /845 (0%). Among the total viral isolates obtained from CSF, the most commonly detected virus was Enterovirus 52 /152 (34%), Adenovirus 38/152 (25%), Epstein-Barr virus 22/152 (15%), Human Herpes virus (6) 17/152 (11 %), Cytomegalovirus 7/ 152 (5%), Herpes simplex virus (1%) 6 /152 (4 %), Human Parechovirus 4 out of 152 (3%), Herpes simplex virus (2) 3 out of 152 (2%), Human Herpes virus (7%) 3/ 152 (2%). Only 216 samples were analyzed for detection of Varicella-Zoster virus and Parvovirus B19 due to unavailability of reagents. The prevalence of these viruses based on the samples analyzed were as follows: Varicella-Zoster 1/216 (1%)
and Parvovirus B19 0/219 (0%) (Table 7).
Figure 2 represents detected virus “human adenovirus”. The Ct value of the positive control is 25. The strongest Ct value was 22 seen in sample 2, which is stronger than positive control, followed by sample 145 and 195 both with Ct values of 24. The highest Ct value was 33 with lower positivity. The negative control did not show any Ct value.
Figure 3 represents the C. neoformans detected. The positive control Ct value was 21, strongest positive sample has Ct value of 33 which is sample 359 and 369. The lower positive has Ct value of 34 in which 18 samples were overlapping at the same Ct value. Negative control does not show any Ct value.
Table 8 shows the use of RT Multiplex PCR; 845 collected CSF samples were analyzed for bacterial meningitis detection, 88 (10%) samples were positive on the following microorganisms: The most detected was S. pneumoniae with 45/88 (51%) followed by Hemophilus influenzae 25/ 88(28%) and Neisseria meningitidis with 18/ 88 (21%).
Figure 4 represents detected bacteria “the Streptococcus pneumoniae”. The positive control Ct value was 24.8; the strongest positive sample has Ct value of 23 which is sample 57. The third positive was known as Streptococcus pneumoniae sample one with Ct value of 26.6. The lower positive has Ct value of 31.8 which was known as Streptococcus pneumoniae sample two. The control negative does not show any Ct value.
Table 9 shows the use of a neonatal kit, 138 CSF samples were collected and analyzed using molecular methods. 17/138 (12%) samples were positive on following microorganisms. The most detected organism was Escherichia coli 15/17 (88%) positive bacteria followed by Listeria monocytogenes 2/17 (12%). No Group B Streptococcus was isolated.
Figure 5 represents the E. coli detected. The strongest Ct value was 7.9. The positive control has Ct value of 26.3. The known sample has Ct value of 26.4. The second strongest Ct value was 10.6 which was sample 559. The third Ct value was 11.6 which was sample 527. The fourth Ct value was 11.9 which was sample 513. The fifth Ct value was 12.4 which was sample 322. The sixth Ct value was 14.8 which was sample 452. The seventh Ct value was 26.4 which was sample 693. The lowest Ct value was 27.5 which was sample 426. The negative control does not show any Ct value.
Figure 6 represents internal control results whereby all Ct values were close together which indicates that the quality of results is quite accurate. This has been used in all samples including negative and positive controls. In some runs, the known sample from accredited external quality control laboratories was included in the runs to ensure the quality of results.
The aim of this study is to evaluate the utility of molecular diagnostic compared with conventional diagnostic methods in detection of etiologic agents of central nervous system infections in Rwanda.
The study was encouraged by data collected from hospitalized patients in four referral hospitals for a period of 4 years from 2009 to 2012, which showed the diagnostic capacity of 9% positive and 91% negative on detection of etiologic agents of central nervous system infections.
The above mentioned results were inadequate for doctors to treat patients with appropriate drugs since some bacteria, fungi and all viruses were not detected using conventional methods. This result will help in early and prompt treatment using appropriate drugs based on isolated micro orgasms by avoiding the prescription of a cocktail of antibiotics, antifungal, or anti-viral drugs.
The developed novel algorithm for molecular diagnostic techniques has been established at National Reference Laboratory, well equipped with high technology equipment and well trained staff. The developed algorithm will help Ministry of Health in Rwanda to come up with policy for quick appropriate diagnosis and treatment of meningoencephalitis. This developed novel algorithm will facilitate Ministry of health to plan for decentralization of molecular diagnostic methods within the laboratory network for better applicability and use, which will greatly assist in patient care, treatment and management at all referral laboratories in Rwanda.
The molecular diagnostic techniques improved diagnostic capacities by detecting 152 viral infections using Real-time Multiplex PCR. This represents 18% detection capacities of viral infections and is in agreement with similar study by (Koskiniemi et al., 2001)which reported molecular methods as being specific, highly sensitive and rapid in detection of viral CNS infections.
This study shows that the diagnostic capacity of etiological agents of CNS infection improved from baseline of 9% diagnostic capacities using routine conventional methods (Ntagwabira et al., 2017), to 40% diagnostic capacities using developed novel algorithm. In this study, the molecular method detected viruses as the most common cause of meningoencephalitis. Enterovirus had the highest viral positivity rate of 34%, followed by Adenovirus 25%, Epstein- Barr virus 15%, Human Herpes virus (6) 11%, and Cytomegalovirus 5%. The lowest positivity rate was Varicella-Zoster 1%. Moreover, based on our findings, the molecular diagnostic method improved diagnostic capacity and therapeutic management of CNS infections by detecting multiple viral infections potentially responsible for mengoencephalitis, as reported similarly by (Quereda et al., 2000).
The detection capacity of bacterial infections improved from 7% using conventional to 22% using molecular diagnostic methods. The usefulness of PCR diagnostic method is supported by previous studies (Issa et al., 2003), which advised the use of PCR diagnostic method in the detection of CNS infection agents. However, the detection capacity of fungal in conventional and molecular methods was the same at 6%.
CONCLUSION AND RECOMMENDATION
This study has shown increased capacity for detection of bacterial and viral causative agents of CNS infections using RT multiplex PCR compared with conventional methods and is recommended for use of molecular diagnostic techniques in diagnosis of CNS infections for better treatment and management of meningoencephalitis suspected patients.
The authors have not declared any conflict of interests.
The authors acknowledge the World Bank through EAPHLNP for funding the research and also appreciate the administrative support provided by the University of Nairobi and Rwanda biomedical centre through SPIU/BIOS/NRL.
REFERENCES
|
Bartt R (2012). Acute Bacterial and Viral Meningitis', CONTINUUM: Lifelong Learning in Neurology 18(6):1255-1270.
|
|
|
|
Baskin HJ, Hedlund G (2007). Neuroimaging of herpesvirus infections in children. Pediatric Radiology 37(10):949-963.
Crossref
|
|
|
|
|
Cho TA, Mckendall RR (2014). Clinical approach to the syndromes of viral encephalitis, myelitis, and meningitis'. Handbook of Clinical Neurology. Elsevier 123:89-121.
|
|
|
|
|
Issa M, Mölling P, Bäckman A, Unemo M, Sulaiman N, Olcén P (2003). PCR of cerebrospinal fluid for diagnosis of bacterial meningitis during meningococcal epidemics; an example from Sudan. Scandinavian journal of infectious diseases 35(10):719-723.
Crossref
|
|
|
|
|
Koskiniemi M, Rantalaiho T, Piiparinen H, von Bonsdorff CH, Färkkilä M, Järvinen A, Kinnunen E, Koskiniemi S, Mannonen L, Muttilainen M, Linnavuori K (2001). Infections of the central nervous system of suspected viral origin: a collaborative study from Finland. Journal of Neurovirology 7(5):400-408.
Crossref
|
|
|
|
|
Logan SAE, MacMahon E (2008). Viral meningitis. British Medical Journal 336(7634):36-40.
Crossref
|
|
|
|
|
Mengistu A, Gaeseb J, Uaaka G, Ndjavera C, Kambyambya K, Indongo L, Kalemeera F, Ntege C, Mabirizi D, Joshi MP, Sagwa E (2013). Antimicrobial sensitivity patterns of cerebrospinal fluid (CSF) isolates in Namibia: implications for empirical antibiotic treatment of meningitis. Journal of Pharmaceutical Policy and Practice 6(1):4.
Crossref
|
|
|
|
|
Minjolle S, Arvieux C, Gautier AL, Jusselin I, Thomas R, Michelet C, Colimon R (2002). Detection of herpesvirus genomes by polymerase chain reaction in cerebrospinal fluid and clinical findings. Journal of Clinical Virology 25:59-70.
Crossref
|
|
|
|
|
Ntagwabira E, Mureithi MW, Muvunyi CM, Masaisa F, Nkeshimana M, Mukagatare I, Thuo D, Jaoko W, Mazarati JB, Anzala O (2017). Challenges in diagnosis of central nervous system infections using conventional method: Need for better approach in Rwanda. African Journal of Microbiology Research 11(35):1379-1385.
|
|
|
|
|
Quereda C, Corral I, Laguna F, Valencia ME, Tenorio A, Echeverria JE, Navas E, Martín-Dávila P, Moreno A, Moreno V, Gonzalez-Lahoz JM (2000). Diagnostic utility of a multiplex herpesvirus PCR assay performed with cerebrospinal fluid from human immunodeficiency virus-infected patients with neurological disorders. Journal of Clinical Microbiology 38(8):3061-3067.
|
|
|
|
|
Richardson AR, Yu Z, Popovic T, Stojiljkovic I (2002). Mutator clones of Neisseria meningitidis in epidemic serogroup A disease. Proceedings of the National Academy of Sciences of the United States of America 99(9):6103-6107.
Crossref
|
|
|
|
|
Taylor WR, Nguyen K, Nguyen D, Nguyen H, Horby P, Nguyen HL, Lien T, Tran G, Tran N, Nguyen HM, Nguyen T (2012). The spectrum of central nervous system infections in an adult referral hospital in Hanoi, Vietnam', PLoS ONE 7(8):e42099.
Crossref
|
|