A novel bacterium pigmented isolate from Caatinga soil was characterized by biochemical and molecular assays, as well as, by rep-polymerase chain reaction (PCR) and 16S rDNA sequencing, and was identified as Serratia marcescens based on 99% of similarity. The identity of the sequences were compared by pairs of critical species of S. marcescens found in National Center for Biotechnology Information (NCBI) with a 96% homology of the isolated species to species database. The wild strain was able to produce biosurfactant (BS) using cassava (Manihot esculenta Crantz) wastewater, with addition of lactose, and corn oil, according to full factorial design 23, at 28°C, and static condition. The net liquid metabolic reduced the surface tension of the water from 70 to 30.60 mN/m during the stationary phase (assay eight constituted by 6.0% cassava wastewater, 1.0% lactose and 7.5% corn oil), and produced emulsifier agent (4.012 UEA) at the same condition. This study identified the pigmented bacterium as new strain of S. marcescens, and showed it has potential to promote both emulsions formation and surface tension reduction in cassava wastewater (6.0%), lactose (1.0%) and corn oil (7.5%) proved as an alternative economical medium for commercial biosurfactant processes.
The semi-arid region of Northeast of Brazil comprises approximately 969.589.4 of km2 (Sousa et al., 2015). The soil composition particularly those under native vegetation for semi-arid is latosols and luvisols under native vegetation for representative sequences from Pernambuco (Sá et al., 2015). This region has a dry tropical deciduous vegetation, constituted by small trees, bushes and grasses, xerophiles, where deciduous plants are the mainly vegetation. It is also characterized by high level of insolation, high temperature, scanty hydric resources and scarce rains, which often cause long periods of drought. In addition, all these factors are usually unfavorable for microbial growth in the soil; however, bacteria constitute the principal group of decomposers responsible for carbon recycling (Solanki and Arora, 2015).
The surfactants in its current use are chemically derived from petroleum; however, interest in microbial surfactants has been applied steadily as surface-active biomolecules that are produced by a variety of microorganisms. The use of low cost substrate can allow a reduction of more than 50% of the final costs of biosurfactant production (Brumano et al., 2016). In this study, cassava (Manihot esculenta Crantz) wastewater effluent was used as substrate. The tubercle has a great economic importance to several cities of Northeastern of Brazil, where its consumption (in terms of carbohydrate content) exceeds those of other crops. However, cassava contain a large amount of cyanogenic glucosides, which can be hydrolyzed to hydrocyanic acid (HCN) by the endogenous enzymes, when the plant tissue is damaged during harvesting, processing or other mechanical processes during the preparations. In this case, the effluent from cassava industrial processing is responsible for the largest environmental impact (Olaifa et al., 2015). The aim of this study was to perform the conventional and molecular identification of the pigmented bacterium isolated from semi-arid soil, as well as to evaluate the potential of the new microorganism to biosurfactant production using cassava wastewater as economic substrate.
Microorganism
The new pigmented bacterium was isolated from Caatinga soil cultured with banana plants, and was deposited in the culture collection of the Catholic University of Pernambuco – UCP. After identification, the strain was registered in World Federation for Culture Collection - WFCC. The strain was maintained on nutrient agar at 5°C.
Substrate
The cassava wastewater was used as media for the production of the biosurfactant, and was obtained from industry food at municipal district of Carnaíba – Pernambuco, and the chemical composition as described by Ponte (1992).
Biochemical and molecular strain identification
The morphological and biochemical characteristic of the isolate bacterium was used for identification tests according to Bergey’s Manual of Systematic Bacteriology (Gorlach-Lira and Coutinho, 2007) and the Manual of Methods for General Bacteriology (Holt et al., 1994).
For identification, the primers FD1 (5'AGAGTTTGATCCTGGCTCAG3’) and RD1 (5'AAGGAGGTGATCCAGCC3') were used to amplify the 16S rDNA fragments. The PCR reaction mixture contained 50 ng genomic DNA (Sambrook and Russell, 2001). Sequences were compared with the ones in the databanks of the National Center for Biotechnology Information, NCBI site (http://www.ncbi.nlm.nih.gov/blast), using the program BlastX (Altschul et al., 1997). The sequences were aligned with the software ClustalW (http://www.ebi.ac.uk) (Thompson et al., 1984). Evolutionary trees for the data set were inferred by the method of Saitou and Nei (1987), and using the neighbor-joining program MEGA (Kumer et al., 1993). The stability of relationships was assessed by performing bootstrap analyses of the neighbor-joining data based on 1.000 resembling.
Medium and culture conditions
The pre-inoculums was prepared by transferring the new strain S. marcescens grown in 100 mL of Luria Bertani (yeast extract 5 g, glucose 5 g, NaCl 5 g per liter), incubated at 28°C, and shacked by orbital shaker at 150 rpm during 17 h. Then, Erlenmeyer’s flasks of 500 mL capacity containing 200 mL of the media according to the factorial design (Table 3) were inoculated with 10% of 106 UFC/mL according to the factorial design (Table 3). The flasks were incubated at 28°C in static condition, during the 72 h. The liquid metabolic free of cells was obtained using filtration through a Millipore membrane with a pore size of 0.22 μm, and were determined pH, surface tension (ST) (mN/m) and emulsification activity (UAE) were determined.
Emulsifier activity test
The metabolic liquid free of cells was added to 2.0 mL of sodium acetate buffer 0.1 M (pH 3), with 1.0 mL of n-hexadecane, the mixture was vortexed for 2 min. After 10 min of rest, the formed emulsions formed were removed with the aid of the Pasteur pipette and transferred and read in spectrophotometer at the wavelength, 540 nm (Cirigliano and Carman, 1984).
Surface tension (ST) measurement
The cell free supernatant was determined in a tensiometer
model Sigma 70 (KSV Instruments LTD - Finland), using the Du Nouy ring method. The reported values are the means of 3 measurements of surface tension (ST) from the media were used as control (Kuyukina et al., 2001).
Experimental factorial design
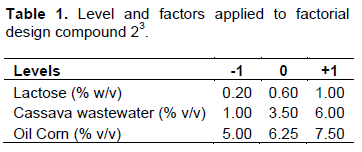
Production of biosurfactant was performed using a full 23 factorial design, augmented with a 4 central points, used to evaluate the impact of 3 independent factors related to lactose, cassava wastewater and corn oil, on the response variable surface tension (ST) and emulsifier activity (Table 1).
Analysis of data was carried out using the analysis of variance (ANOVA). The results were analyzed then examined to determine the main effects of all factors. The ANOVA was carried out to determine which factors are statistically significant. The data analyses and graphs, and all calculations were performed using the Statistical 7.0 software package, and significance of the results was tested at the (p≤0.05).
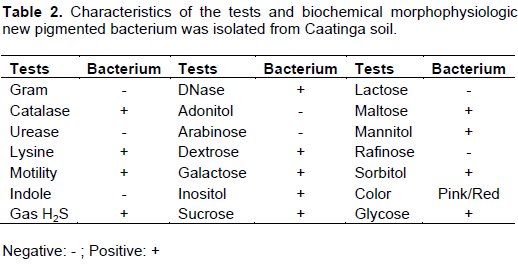
The Gram negative bacterium isolated from Caatinga banana tree soil showed red colonies on LB medium at 28°C during 24 h. The Rugai-Lysine test was employed (Pessoa and Silva, 1972) and catalase test showed positive reaction. The motility of gas production, indole, urease and sucrose showed negative reactions. The results identified the strain as Enterobacteriacea family, genus Serratia (Table 2).
The optimum growth conditions of Serratia were observed in pH 9 at temperatures of 20 to 30°C (Khanafari et al., 2006). The results obtained agree with those reported by Dundar et al. (2009). This method is widely used in bacterial systems. Apart from the morphology, staining can reveal certain properties of the bacteria which are essential for classifying it. Under this method, bacteria are classified into two groups: Gram positive and negative, the isolate examined was a Gram negative bacillus, which is alcohol acid resistant.
The sample also showed red-pink color when grown in Luria Bertani media at 28°C for 24 h. The Rugai-Lysine test was employed (Pessoa and Silva, 1972) and showed a positive reaction for lysine, motility and the production of gas, and a negative reaction for indole, urease and sucrose. Therefore, with this method, the examined strains can be identified as a genus Serratia sp. of the Enterobacteriacea species. The catalase test was positive for the sample, so the results of the morphological and biochemical analysis of the isolate can identify it as an Enterobacteriacea species of the genus Serratia marcescens, according to Holt et al. (1994).
The bacteria of the species S. marcescens may be identified in the laboratory through Gram-staining, although sometimes they can be observed without the use of laboratory equipment since at room temperature they display pink to red pigmentation. S. marcescens displays an intense red pigmentation, prodigiosin by age of the colonies (Rastegari and Karbalaei-Heidari, 2016). To confirm the morphological and biochemical tests, molecular tests were also used. The DNA isolate was subjected to PCR reactions with primers specific for 16S rRNA region, which resulted in the amplification of a fragment of approximately 1200 pb (Figure 1). Genes of 16S rDNA have long been the standard for systematic molecular studies, as well as for the rapidly expanding fields of molecular diagnostics and microbial ecology. These genes are universal multiple copies of 16S rDNA genes of the genome; thus making it abundant, easily detectable. Primers were designed to amplify fragments specific to the analysis of sequence. Gaps in 16S rRNA as a target for detection and identification are related to the fact, and they are often insufficient to discriminate the sequence information within the target 16S rRNA in order to distinguish among closely related species and strains (Ma et al., 2016).
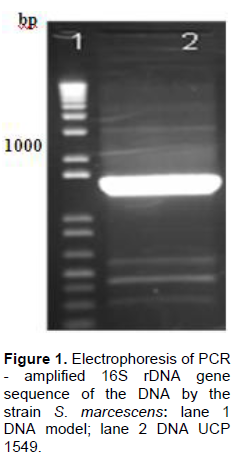
The isolated DNA was subjected to PCR reactions with specific primers for 16S rRNA, which resulted in the amplification of a fragment of approximately 1200 bp (Figure 1). The 16S rDNA have been used as standard for molecular systematic studies as well as the rapid expansion of the areas of molecular diagnostics and microbial ecology (Vierheilig et al., 2015).
The complete nucleotide sequence of isolate by 16S rDNA gene, was compared by blast software specifically designed by the NCBI GenBank (BLAST/bl2seq) (GenBank). The identity of the sequences were compared by pairs of critical species S. marcescens found in NCBI/National Center for Biotechnology Information, a 96% homology of the isolated species to species database of NCBI was observed. The construction of the phylogenetic tree (Figure 2), in conjunction with the BLAST search, was gaining immense importance among researchers. This method relates the strain in question with those exhibiting a sequence homology of DNA and helps to overcome the possibility of errors by performing statistical analysis, such as testing the boot (Weisburg et al., 1991).
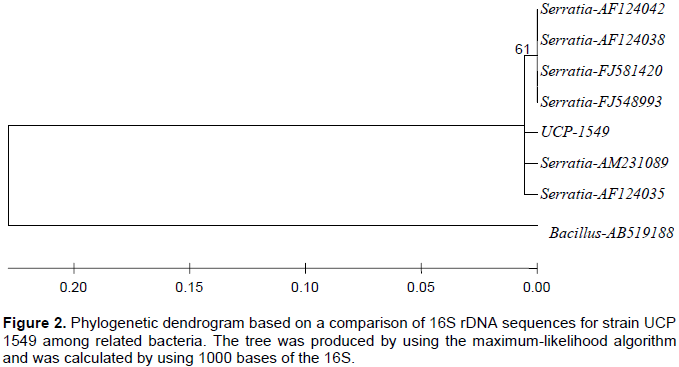
Two bodies can only be compared if they show similarity at least 80% in the primary sequences of their DNA. Indeed, hybrids showing 90-95% similarity in their sequences of nucleotide bases of DNA hybridization, showed 50% DNA (Weisburg et al., 1991). Most studies used this method, organisms that have a high homology (above 50%) have similar phenotypic characteristics. The correlation between phenotypic and genotypic properties led to the proposal that organisms with prokaryotic DNA homology values of less than 70% are considered as belonging to the same genomic species. The analysis of the 16S rDNA sequences confirmed the strain catalogued as UCP 1549 was closely related to S. marcescens based on 99% of similarity in their 16S rDNA sequences. The results indicate that amplified 16S rDNA analysis is a sample and reliable tool for the identification of bacterial species according to the literature (Zhu et al., 2007; Tariq and Prabakaran, 2011).
Cassava wastewater was a nutrient source for biosurfactant production by S. marcescens and the use of agroindustrial substrate manipueira could reduce the economic process of production. On the other hand, cassava processing is generally considered to contribute significantly to environmental pollution and aesthetic nuisance. In this study, the identified bacterium, S. marcescens demonstrated ability for a very fast growth and to convert cassava wastewater, lactose and corn oil are important nutrients to biosurfactant production detected by surface tension, emulsifier activity using a factorial design, after 72 h of growth, in the static condition. The best results observed were a significant surface reduction tension of water (70 mN/m to 30.60 mN/m) in condition 8 of the factorial design, with greater concentration of cassava wastewater (6.0%), lower level of lactose (1%), and higher level of corn oil (7.5%). At the same time, higher emulsifier activity in the assay 4 (cassava wastewater 6.0%, lower level of lactose 1%, and higher level of corn oil 5%) was observed (Table 3). According to the Pareto diagram (Figure 3A), the independent corn oil and lactose and interaction lactose/corn oil favored positively the reduction of surface tension; however, the interaction lactose/cassava even considered significant at a confidence level within 5 % (p <0.05) and did not contribute to the reduction but was not statistically representative. The emulsification activity according to the Pareto diagram (Figure 3B) showed that cassava had no effect on emulsifying activity and corn oil as well as the interactions between lactose/cassava, lactose/corn oil, which was statistically more significant influence the emulsification activity. However, the emulsification activity was very promising for all conditions of the factorial design, with the highest emulsification activity in test 4 (4.012 UAE), and lowest in the assay 8 (UAE 1.798) (Table 2).
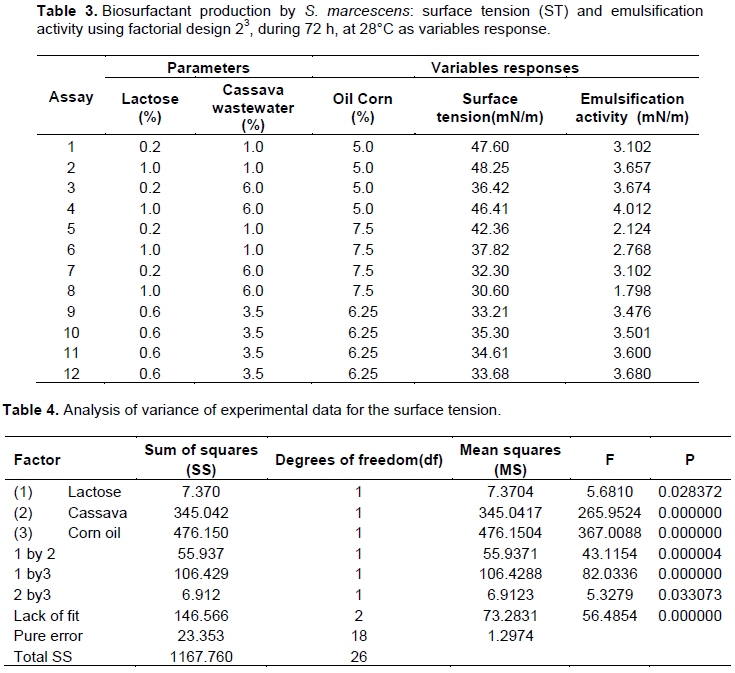
A study was carried out using Serratia sp with a reduction of the surface tension to 27.8 mN/m, after 72 h of cultivation (Montero-Rodríguez et al., 2015). Similar results with S. marcescens was observed by Ferraz et al. (2002) which demonstrated the ability for reduction of the surface tension to 29.75 mN/m by, but using orbital shaker. The obtained statistical model explained 85% of data variation and provide accreditations established of the experimental model. The Pareto chart (Figure 3A) reinforces the prediction model established by ANOVA. The statistical model of accuracy of surface tension as a function of the parameters was obtained by the following equation:
TS = 49.84 + 22.36 [Lactose] – 1.36 [Cassava] – 0.43 [Corn oil] + 1.53[lactose] [cassava] – 4.21[corn oil] [lactose] – 0.17 [cassava] [corn oil].
Araújo et al. (2009) described that the previous results of biosurfactant production by S. marcescens using as substrate lactose, corn oil and corn step liquor in substitution of cassava waste water and the reduction of the surface tension was 33.35 mN/m. Experimental factorial design conducted with Serratia sp. showed a reduction of the surface tension of the medium from 67.8 to 34.4 mN/m after 96 h of cultivation (Cunha et al., 2004).
The stability of the biosurfactant was evaluated by the pH, the saline concentration and the temperature. Several studies described the of biosurfactants production optimization by culture media of low cost associated with industrial residue as insoluble substrates (Silva et al., 2010; Joshi et al., 2013; Ebadipour et al., 2016).
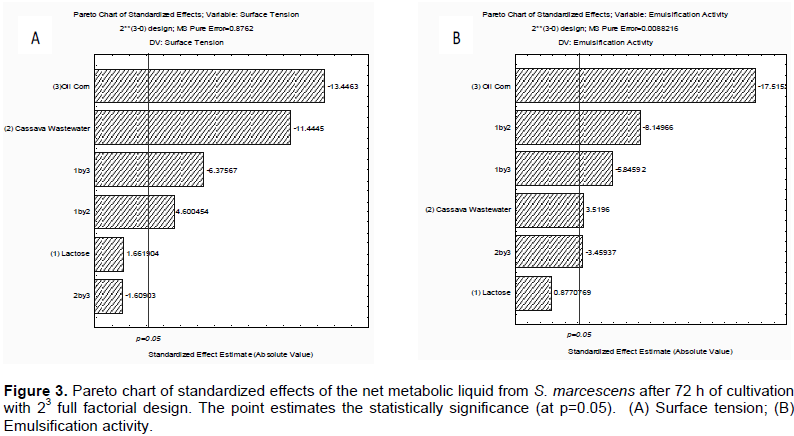
The biosurfactant produced by S. marcescens in the best condition of factorial design (assay 8) evaluated by surface tension of cell-free metabolic net showed stability under all different pH, NaCl concentrations and temperature.
The stability of the produced biosurfactant produced was influenced in reducing surface tension by acid pH, salt concentration up to 6%, and the temperature 100°C (Figures 4A, B and C), respectively. However, there was no significant difference in the variation of surface tension, indicating its potential application in aquatic environments, because it is independent of the used concentration of NaCl and the biosurfactant remained effective in reducing the surface tension (Figure 4B). These results of the stability of the biosurfactant to 10% NaCl concentration, considering that conventional surfactants are inactivated by NaCl concentrations of 2 and 3%, as described by Rufino et al. (2007). The response surface graphs lactose/corn oil (Figure 5B), cassava/lactose (Figure 5A), have significant interactions, that is, cannot be explained by variations alone, because their curves were not parallel. Figure 5C showed the curves of levels of character parallel, and surface response indicated considerable interaction between the used media.
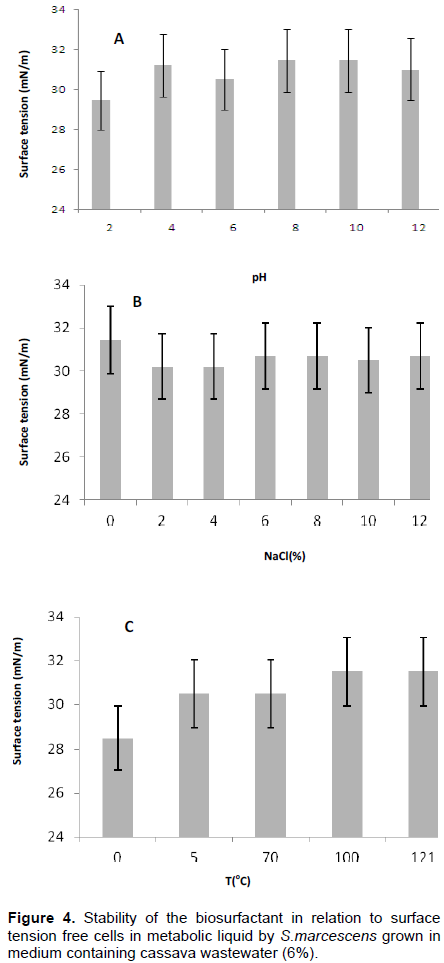
The experimental data showed the analysis considering to the characteristics: the main significant effects and interactions among the means employed, according to the values F (Fischer constant) and P-(value confidence interval< 0.05). The pure error or experimental error value was considerably reduced; 56.5 times lower than the error on the fit (experimental data).
The conventional taxonomy and 16S rDNA gene determination confirmed the identification of the pigmented bacterium, as new strain of S. marcescens. In addition, the DNA homology has shown the comparison among all types of prokaryotic microorganisms, and confirmed the identification regardless of their conditions of growth or metabolism and confirmed the identification. The new strain of S. marcescens showed ability to grow on agroindustrial substrate, cassava wastewater and both productions, a potent tensioactive and emulsification agents in the static condition, in the economic and low cost media. The biosurfactant was stable when tested at different pH, temperature and concentrations of NaCl measured by surface tension. The statistic model to tensio-active agent indicated better interactions in the independents variables. The biosurfactant produced by S. marcescens, can be used for bioremediation process of environmental pollution with hydrophobic substances.