ABSTRACT
Lysobacter. spp. are considered as important biocontrol bacteria, due to their antagonistic activity against many pathogenic fungi, bacteria and nematodes. Rep-PCR was used to analyze the genetic diversity of 12 Lysobacter strains. These strains included Lysobacter antibioticus (HY, 13-1, 13-6, 6-B-1, 13-B-1, 6-T'-4, LJ6-3, LJ6-4 and LR9-3), Lysobacter enzymogenes (DH3, 1-T-1-4) and Lysobacter capsici (LG18) isolate from different regions in Yunnan province of China. Rep-PCR was performed using DNA amplification with primers based on short bacterial repetitive elements (ERIC, BOX, IS1113 and J3). The genetic diversity was analyzed through rep-PCR, molecular fingerprint clustering analysis and UPGMA to construct phylogenetic tree. The results show that when the genetic distance was 0.59, IS1113-PCR could cluster the Lysobacter strains as 3 species: L. antibioticus, L. enzymogenes and L. capsici. The 3 species had obvious differences between each other and the rep-PCR technique could be used to detect genetic variation between different Lysobacter strains, identification and strain classification.
Key words: Lysobacter, Rep-PCR, genetic diversity.
With the development of molecular biology technology, in recent years, a variety of techniques based on PCR has been widely used in genetic diversity analysis of pathogenic fungi and bacteria, especially the appearance and use of rep-PCR, it made the biodiversity research more rapid, convenience and economy (Xiang et al., 2010; Li et al., 1999). Due to the repetitive DNA sequence only in prokaryotes, but not in the eukaryotic chromosomes, rep-PCR technology can specify amplification prokaryotes DNA, and avoid the influence of eukaryotic gene, so it is especially suitable for related eukaryotes prokaryotic symbiotic bacteria and pathogenic bacteria of genetic diversity research (Li and Zhi, 2006). This technique use the highly conserved short repeat sequences in fungal or bacterial genomic DNA as target sequences of primers to amplify through PCR. Through agarose gel electro-phoresis separating DNA fragments with different sizes, each species or strains can produce specific DNA fingerprints. Rep-PCR was used to analyze the genetic diversity of Xanthomonas strains and results indicated rep-PCR was a useful tool for detecting genetic variation among strains of X.o.pv.oryzicola and identi-fication of strains as well as classification studies (Ji et al., 2002).. REP-PCR and BOX-PCR were used to study the genetic diversity of Ralstonia solanacearum and also indicated that this tool was scientific and effective (Li et al.,2011).
Lysobacter. spp. belong to the Xanthomonadaceae family, produce yellow to brownish black pigment, rod-shaped cells, rounded ends, no flagella, the edge of the colony is clear and smooth, have the ability to slip (Ji, 2011). Lysobacter are widely distributed in nature and found in soil, rivers, sewers, and other extreme environment (Christensen and Cook, 1978). Lysobacter species are recognized as new bacterial predators with the arsenal of bioactive small molecules, the biosynthetic mechanisms and biosynthetic genes for cyclodepsipeptide lysobactin, cyclic lipodepsipeptides, cephem-type β-lactams and polycyclic tetramate macrolactams which make them as biocontrol agents and promising drug producers (Xie et al., 2012). In 1978, Christensen named 4 species of this genus of bacteria: Lysobacter enzymogenes, Lysobacter antibioticus, Lysobacter brunescens and Lysobacter gummosus (Christensen and Cook, 1978). With the development of bacterial taxonomy technology and a large number of professional database updating, this bacteria was reclassified from Stenotrophomonas to Lysobacter spp. (Sullivan et al., 2003; Islam et al., 2005). Forepart, the research mainly focused on its activity of various extracellular enzymes, they act on the pathogen cell wall and other targets, causing lysis and inhibition (Reichenbach, 2006). Lysobacter spp. are considered as new microbial pesticides, and produce extracellular enzymes and antibiotics that can inhibit the occurrence of several plant diseases. Anovel antifungal compound (HSAF, 1) was isolated from L. enzymogenes C3 and can disrupt sphingolipids important to the polarized growth of filamentous fungi, thus, L. enzymogenes C3 was used in the biological control of fungal diseases of plants (Lou et al., 2011). L enzymogenes OH11 secretes chitinases that hydrolyzed the pathogenic fungal cell wall contributing to suppression of proliferation (Postma et al., 2009); Lysobacter sp. strain sbk88 and L. enzymogenes 3.1t8 can produce antibiotics that are involved in suppression of Pythium aphanidermatum and Aphanomyces cochlioides (Kato et al., 1998). L. enzymogenes OH11 can control Pseudomonas solanacearum (Jiang et al., 2005) and L. antibioticus 13-1 was shown to control Xanthomonas oryzae pv. Oryzae and Erwinia carotovora subsp. carotovor , Ecc postulated by niche exclusion by colonizing the crop rhizosphere and competition for nutrients (Wei et al., 2014; Zhang et al., 2011; Wu et al., 2010).
Most studies on Lysobacter in recent years were all focused on its biological control, gene cloning and protein (Qian, 2009; Nian, 2015; Liu, 2012). Irene de Bruijn et al. (2015) studied genomics and metabolic profiling of Lysobacter in 2005 (de Bruijn et al., 2015). rep-PCR was utilized to analysis 12 strains of Lysobacter in order to investigate the diversity of this important biocontrol bacterial genome and lay the foundation for application.
Test materials and instruments
Test strains and culture medium
L. antibioticus (HY, 13-1, 13-6, 6-B-1, 13-B-1, 6-T'-4, LJ6-3, LJ6-4, LR9-3), L. enzymogenes (DH3, 1-T-1-4) and L. capsici (LG18) were isolated from different regions in Yunnan province of China.
NA medium: sucrose 10 g, peptone 5 g, beef extract 3 g, yeast extract 1 g, agar 15 to 18 g, water after dissolving to volume 1000 mL, adjusted to a pH of 7.0, high-pressure sterilized.
R2A medium: peptone 0.5 g, acid hydrolyzed casein 0.5 g, glucose 0.5 g, amylogen 0.5 g, yeast extract 0.5 g, C3H3NaO3 0.3 g, K2HPO4·3H2O 0.3 g, MgSO4·7H2O 0.005 g, agar 15 g, dissolved in water after constant volume to 1000 mL, adjusted to a pH of 7.0, high-pressure sterilized.
LB liquid medium: peptone 10 g, yeast extract 5 g, 10 g sodium chloride, dissolved in water after constant volume to 1000 mL, adjusted to a pH of 7.0, high-pressure sterilized.
Major instruments and reagents
Super clean bench, Constant temperature shaking table, Small high-speed centrifuge, Thermostatic water bath, PCR instrument (Takara), High-pressure steam sterilization pot, Image master UV imaging system , Bacterial Genomic DNA Extraction Kit (Beijing Taike Biotechnology Co., Ltd. 50 times), PCR primers by Takara Biotechnology (Dalian) Co., Ltd. synthesis; Takara SYBR premix ExTaq (perfect real time kit purchased from Takara Biotechnology (Dalian) Co., Ltd.
Isolation and identification of Lysobacter from soil samples
Dilution coated plate separation method was used (Wang et al., 2007), the separation is mainly based on the color and shape of colonies to screen Lysobacter in NA and R2A plates, then 16S rDNA of bacterial universal PCR amplification primers 27F (AGAGTTTGATCCTGGCTCAG)/1492R (GGTTACCTTGTTACGA CTT) are used. PCR reaction system (25 μL): 2 x EasyTaq PCR Supermix 12 μL, primer 27F 1 μL, primer 1492R (10 μmol / L) 1 μL, DNA template (50 ng/ μL) 1 μL, ddH2O complement 25 µL. The initial denaturing temperature was 95°C for 5 min, followed by 30 cycles of 94°C for 30 s, 50°C for 30 s, and 72°C for 1.5 min, with a final extension at 72°C for 10 min. Amplification products was by 1.0% agarose gel electrophoresis to recover. After purification, 16S rDNA fragments sequencing by Shanghai Biological Engineering Co., Ltd. sequencing primers are 27F and 1492R. Sequencing results blast in GenBank were then obtained with high similar sequence. A total of 12 strains of bacteria were isolated, purified and identified from the collected soil samples (Table 1).
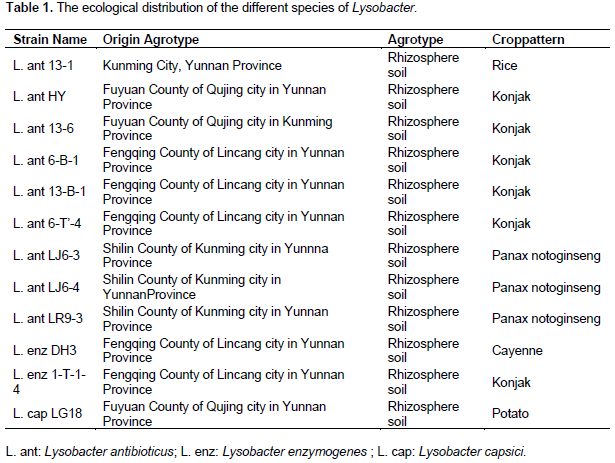
Rep-PCR and genetic diversity analysis of Lysobacter
Lysobacter genome DNA extraction (referring to Beijing Taike Biotechnology Co., Ltd. bacterial genomic DNA extraction kit), and then rep-PCR was done.
DNA was used to perform rep-PCR with ERIC primers using the manufacturer’s protocol. The primer pair Eric1R: 5´-ATGTAAGCTCCTGGGGATTCAC-3´, Eric2: 5'-AAGTAAGTGACTGGGGTGAGCG-3´ was used and composed by the Shanghai born industry and Biological Engineering Co., Ltd. synthetic. PCR reaction system: 10 x PCR buffer, 50 ng of DNA template, 400 µM dNTP, primer F 10 pmol, primer R 10 pmol, 2 U Taq enzyme, ddH2O complement 25 µL. The initial denaturing temperature was 95°C for 7 min, followed by 35 cycles of 94°C for 1 min, 52°C for 1 min, and 65°C for 8 min, with a final extension at 65°C for 15 min.
The primer BOX: 5´-CTACGGCAAGGCGACGCTGACG-3´ was used and composed by the Shanghai born industry and Biological Engineering Co., Ltd. synthetic. PCR reaction system: 10 × PCR buffer, 50 ng of DNA template, 400 µM dNTP, primer F 10 pmol, primer R 10 pmol, 2 U Taq enzyme, ddH2O complement 25 µL. The initial denaturing temperature was 95°C for 7 min, followed by 35 cycles of 94°C for 1 min, 53°C for 1 min, and 65°C for 8 min, with a final extension at 65°C for 15 min.
The primer J3: 5´-GCTCAGGTCAGGTCGCCTGG-3´ was used and composed by the Shanghai born industry and Biological Engineering Co., Ltd. synthetic. PCR reaction system: 10 × PCR buffer, 50 ng of DNA template, 400 µM dNTP, primer F 10 pmol, primer R 10 pmol, 2 U Taq enzyme, ddH2O complement 25 µL. The initial denaturing temperature was 95°C for 7 min, followed by 35 cycles of 94°C for 1 min, 68°C for 1 min, and 65°C for 8 min, with a final extension at 65°C for 15 min.
The primer pair IS1113: TX1: 5´-TGTAGTGGACCTTCGAA-3´, TX2: 5´-ACGAGCGATTGATCAGG-3´ was used and composed by the Shanghai born industry and Biological Engineering Co., Ltd. synthetic. PCR reaction system: 10 x PCR buffer, 50 ng of DNA template, 400 µM dNTP, primer F 10 pmol, primer R 10 pmol, 2 U Taq enzyme and ddH2O complement 25 µL. The initial denaturing temperature was 95°C for 7 min, followed by 35 cycles of 94°C for 1 min, 50°C for 1 min, and 65°C for 8 min, with a final extension at 65°C for 15 min.
The end of amplification, 8 µL PCR product was taken in 0.5 * TAE buffer through the concentration of 1.5% agarosegel electrophoresis, the voltage drop to 100 V, electrophoresis was done for 5-6 h, after the completion of the electrophoresis, using ethidium bromide staining, decolorization, in gel imaging system amd the collection and preservation of image was done.
Approximately 25 mL of the post amplification reactions were separated by electrophoresis in a 1.5% agarose gel in TAE buffer, stained with an ethidium bromide solution (10 mg/mL), decolorization and photo documented under UV light. An optimized comparative study of the diversity of the obtained banding patterns showed the presence or absence of repetitive elements using photographic images.
Rep-PCR amplified products of the gel map to read the band, have band (mark "1"), no band (mark "0"), using software NTSYS for cluster analysis, and using unweighted pair group method with arithmetic mean (UPGMA) for molecular fingerprint clustering analysis and constructing system tree pattern.
Analysis of rep-PCR fingerprint banding patterns
The Box PCR showed 11 different molecular fingerprints with bands ranging from 250 to 7000 bp for the 12 isolates of L. antibioticus (HY, 13-1, 13-6, 6-B-1, 13-B-1, 6-T'-4, LJ6-3, LJ6-4, LR9-3), L. enzymogenes (DH3, 1-T-1-4) and L.capsici (LG18) (Figure 1).
The Eric PCR showed 17 different molecular fingerprints with bands ranging from 250 to 7000 bp for the 12 isolates of L. antibioticus (HY, 13-1, 13-6, 6-B-1, 13-B-1, 6-T'-4, LJ6-3, LJ6-4 and LR9-3), L. enzymogenes (DH3, 1-T-1-4) and L.capsici (LG18) (Figure 2).
The IS1113 PCR showed 5-15 different molecular fingerprints with bands ranging from 250 to 7000 bp for the 12 isolates L. antibioticus (HY, 13-1, 13-6, 6-B-1, 13-B-1, 6-T'-4, LJ6-3, LJ6-4, LR9-3), L. enzymogenes (DH3, 1-T-1-4) and L.capsici (LG18) (Figure 3).
The J3 PCR showed 8-16 different molecular fingerprints with bands ranging from 250 to 7000 bp for the 12 isolates of L. antibioticus (HY, 13-1, 13-6, 6-B-1, 13-B-1, 6-T'-4, LJ6-3, LJ6-4 and LR9-3), L. enzymogenes (DH3, 1-T-1-4) and L. capsici (LG18) (Figure 4).
Results of PCR fingerprint analysis
UPGMA cluster analysis was performed on the DNA fingerprints and showed that only primer IS1113 can distinguish L. antibioticus (r1-9), L. enzymogenes(r10-11) and L. capsici L.capsici (r12) as three groups (Figure 5). According to different geographical locations, we can reclassified L. antibioticus HY and L. antibioticus 13-6 both from Fuyuan County of Yunnan Province as the first group, L. antibioticus 13-1 from Kunming City of Yunnan Province as the second group, L. antibioticus 6-B-1, 13-B-1 and 6-T'-4 from Fengqing County of Lincang City in Yunnan Province as the third group, L. antibioticus LJ6-3 and LJ6-4 from Shilin County of Yunnan Province as the fourth group, L. antibioticus LR9-3 from Shilin County of Kunming city in Yunnan Province as the fifth group, L. enzymogenes 1-T-1-4 and DH3 both from Fengqing County of Lincang city in Yunnan Province as the sixth group and L. capsici LG18 from Fuyuan County of Qujing city in Yunnan Province as the seventh group (Table 1). The results confirmed that as compared to the similarity between species, the similarity of the isolates within in the species was higher (Table 2).
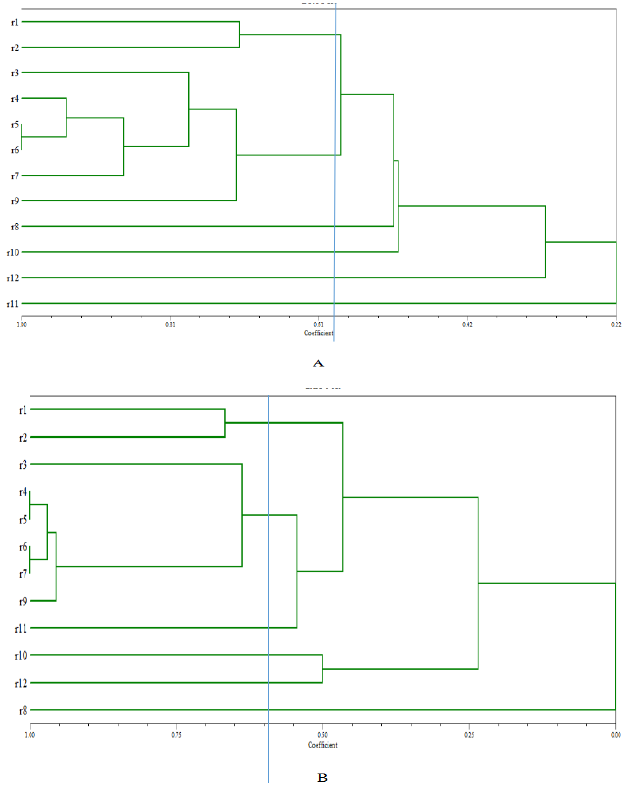
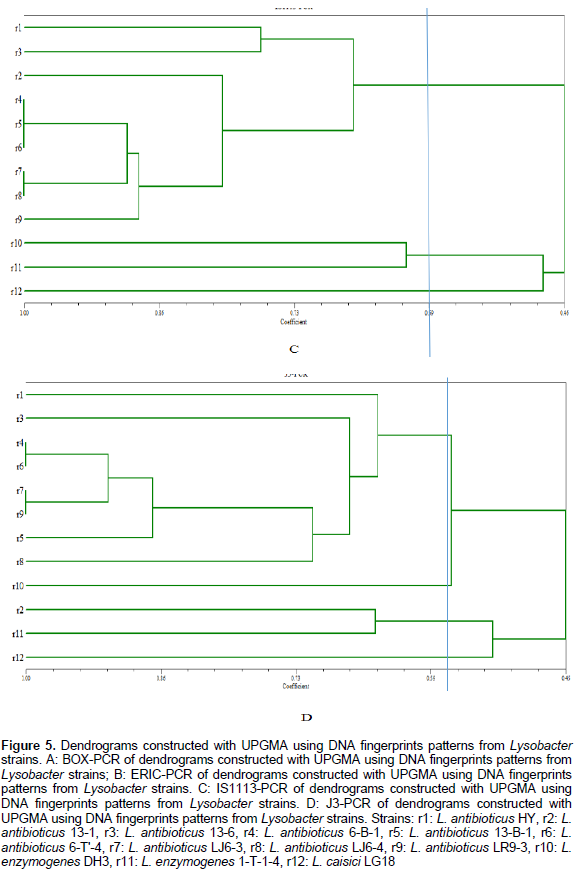
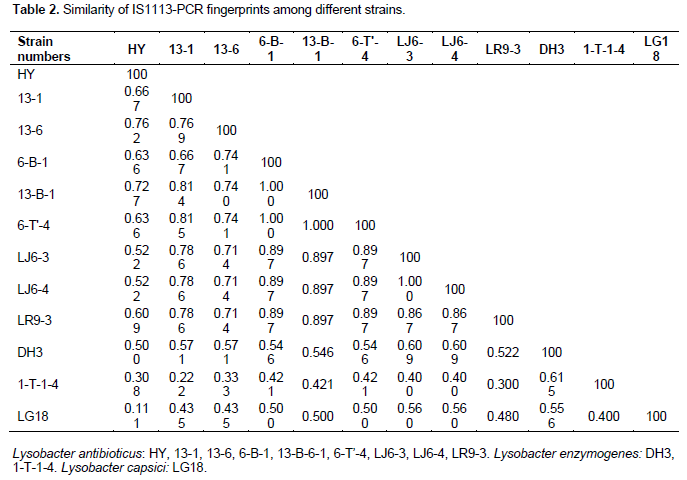
The BOX, ERIC, IS1113 and J3 repeat sequences are distributed widely in three species of Lysobacter genome DNA, and confirmed rep-PCR technology can distinguish different species and used for the determination of Lysobacter group genetic diversity. Primers of BOX, ERIC, IS1113 and J3 are used for PCR, fingerprint and clustering analysis, results show that when genetic distance was 0.59 only IS1113 primer can successfully distinguish L. antibioticus, L. enzymogenes and L. capsici. IS1113 can be used for analysis of hereditary changes in Lysobacter groups, except a few strains, which can be clearly divided into three main genetic clusters. Lysobcater have different structure, activity and biosynthetic pathways, thus the control of pathogenic bacteria is different. So we can classify species quickly through rep-PCR and then use specific biocontrol bacteria to control plant diseases.
It is the first time to apply rep-PCR to study the diversity of Lysobacter genome. The method of identifying Lysobacter usually uses 16S rDNA of bacterial universal primers to PCR and then amplified products are sequenced, with sequencing results blast in GenBank and then high similar sequence was obtained. This method is time consuming, inconvenient and the results are not accurate because the sequencing results may be inaccurate. Thus, rep-PCR was adopted, this technology has advantages of fast, handy and economic, and dispense with specific probe and southern hybridization. Due to the characteristics of electrophoresis, rep-PCR with strain level spectrum can be used for strain identification and can reflect the differences of genome of close genetic relationship between strains, but the disadvantage is that it does not reflect the differences in the plasmid DNA, and rep-PCR is affected by many factors, such as: primers from different sources or different batches, DNA polymerase or PCR instrument of different types, to an extent, these factors restrict the technology application (Laguerre et al., 1996). In spite of this, under certain experimental conditions, rep-PCR is an important and effective technology for strains identification and clustering. In the later work, Multilocus Sequence Analysis (MLSA) and Restriction Fragment Length Polymorphism Analysis (RFLP) can be used to further research on the origin of their diversity, these will be helpful to further uncover the origin, evolution and phylogeny, and provide important information and scientific basis in the production or use of Lysobacter for disease prevention and economic crops treatment.
The authors declare that there is no conflict of interest.
This project was funded by National Natural Fund (31360002, 31460458), The Ministry of Agriculture Public Welfare Industry Special (201303015), Province Key New Product Plan (2014BB005) and Yunnan University Innovation Team IRTSTYN (2014-22).
REFERENCES
|
Christensen P, Cook F D(1978). Lysobacter, a new genus of nonfruiting gliding bacteria with high base ratio. Int. J. Syst. Bacteriol. 28:367-398.
Crossref
|
|
|
|
de Bruijn I, Cheng X, de Jager V, Expósito RG, Watrous J, Patel N, Postma J, Dorrestein PC, Kobayashi D, Raaijmakers JM (2015). Comparative genomics and metabolic profiling of the genus Lysobacter. BMC Genomics 16:991.
Crossref
|
|
|
|
|
Islam MT, Hashidoko Y, Deora A, Ito T, Tahara S (2005). Suppression of damping-off disease in host plants by the rhizoplane bacterium Lysobacter sp. strain SB-K88 is linked to plant colonization and antibiosis against soilborne Peronosporomycetes. Appl. Environ. Microbiol. 71:3786-3796.
Crossref
|
|
|
|
|
JI Guang-hai (2011). Advances in the Study on Lysobacter spp. Bacteria and Their Effects on Biological Control of Plant Diseases. J. Yunnan Agric. Univ. 26(1):124-130.
|
|
|
|
|
JI Guang-hai, XU Zhi-gang, ZHANG Shi-guang (2002). Preliminary Analysis of Genetic Diversity of Xanthomonas oryzae pv.oryzicola and Xanthomonas leersiae Stains in China by Rep-PCR. Acta Phytopathol. Sin. 32(1):26-32.
|
|
|
|
|
Jiang Ying-hua, Hu Bai-shi, Liu Feng-quan (2005). Selection and Identification of Antagonistic Bacteria against Soil-borne Plant Pathogens. Chin. J. Biol. Control 21(4):260-264.
|
|
|
|
|
Kato A, Nakaya S, Kokubo N, Aiba Y, Ohashi Y, Hirata H, Fujii K, Harada KI (1998). A new anti-MRSA antibiotic complex WAP-8294A. I. Taxonomy, isolation and biological activities. J. Antibiot. 51(10):929-935.
Crossref
|
|
|
|
|
Laguerre G, Mavingui P, Allard MR, Charnay MP, Louvrier P, Mazurier SI, Rigottier-Gois L, Amarger N (1996). Typing of rhizobia by PCR DNA fingerprinting and PCR-restriction fragment length polymorphism analysis of chromosomal and symbiotic gene regions: application to Rhizobium leguminosarum and its different biovars. Appl. Environ. Microbiol. 62(6):2029-2036.
|
|
|
|
|
Li Hai-yun, Che Jian-mei, Liu Bo, XueFang Z, RongFeng X (2011). Genetic Diversity Analysis of Ralstonia solanacearum Based on BOX-PCR and REP-PCR. J. Agric. Biotechnol. 19(6):1099-1109.
|
|
|
|
|
Li Jie, Xiong Zhi (2006). Studies on Genomic Diversity of Bacteria by rep-PCR. J. Southwest For. Coll. 26(6):1-3.
|
|
|
|
|
Li J, Xu L, Fan H, Li L, Ge C, Yang S (1999). Use rep-PCR for Study the Diversity of Chinese Peanut Nodule Bacteria. J. Microbiol. 39(4):296-303.
|
|
|
|
|
Liu Yi-ru (2012). Identification and Function Characterization of Quorum Sensing System in Lysobacter enzymogenes. Nanjing Agricultural University pp. 1-69.
|
|
|
|
|
Lou L, Qian G, Xie Y, Hang J, Chen H, Zaleta-Rivera K, Li Y, Shen Y, Dussault PH, Liu F, Du L (2011). Biosynthesis of HSAF, a tetramic acid-containing macrolactam from Lysobacter enzymogenes. J. Am. Chem. Soc.133:643-645.
Crossref
|
|
|
|
|
Nian Mei-yu (2015). Identification of potential antibacterial activity proteins on Lysobacter sp.SNNU513. Shanxi Normal University. pp. 1-63.
|
|
|
|
|
Postma J, Stevens LH, Wiegers GL, Davelaar E, Nijhuis EH (2009). Biological control of Pythium aphanidermatum in cucumber with a combined application of Lysobacter enzymogenes strain 3.1T8 and chitosan. Biol. Control 18:301-309.
Crossref
|
|
|
|
|
Qian Guo-liang (2009). Cloning and Characterization of Biocontrol Related Genes and Construction of Engineering Strain in Lysobacter enzymogenes OH11. Nanjing Agricultural University pp. 1-185.
|
|
|
|
|
Reichenbach H (2006). The genus Lysobacter. Prokaryotes 6:939-957.
Crossref
|
|
|
|
|
Sullivan RF, Holtman MA, Zylstra GJ, White JF Jr., Kobayashi DY (2003). Taxo- nomicpositioning of two biological control agents for plant diseases as Lysobacter enzymogenes based on phylogenetic analysis of 16S rDNA, fatty acid composition and phenotypic characteristics. J. Appl. Microbiol. 94:1079-1086.
Crossref
|
|
|
|
|
Wang P, Yang ZW, Hu HT, Wang KM, Zhang F (2007).Elementary Research on the Screening of Antagonisms and Their Antibiotic Activities Against the Phytopathogens. Hubei Agric. Sci. 46(2):236-238.
|
|
|
|
|
Wei Lan-Fang, Zhou Li-Hong, JI Guang-Hai, Wang Yong-Ji, WANG Shao-Xue (2014). Control of rice bacterial leaf blight by antibacterial substances from Lysobacter antibioticus strain 13-1. Microbiology 41(2):274-280.
|
|
|
|
|
Wu Ya-peng, JI Guang-hai, Chen Yun-lan, Lu Jun, Dong Kun (2010). Biocontrol Effect and Mechanisms of Lysobacter antibioticus 13-1 against Soft Rot Pathogen of Amorphophallus konjac. Chin. J. Biol. Control 2:193-199.
|
|
|
|
|
Xiang Ming-jie, Liu Jin-yan, Ni Pei-hua, Zhang Hua, Ni Yu-xing (2010). Repeat Sequence PCR (rep-PCR) Fingerprint Analysis in the Identification of Pathogenic Fungi and Classification. Chin. J. Mycol. 5(6):376-380.
|
|
|
|
|
Xie Y, Wright S, Shen Y, Du L (2012). Bioactive natural products from Lysobacter. Nat. Prod. Rep. 29:1277-1287.
Crossref
|
|
|
|
|
Zhang Li-hui, Wang Yong-ji, Liao Lin, Ji Guang-hai (2011). Biocontrol effect of Lysobacter antibioticus 06−4 on soft rot pathogen of Amorphophallus konjac its mechanism. J. Hunan Agric. Univ. (Natural Sciences) 3:286-289.
Crossref
|
|