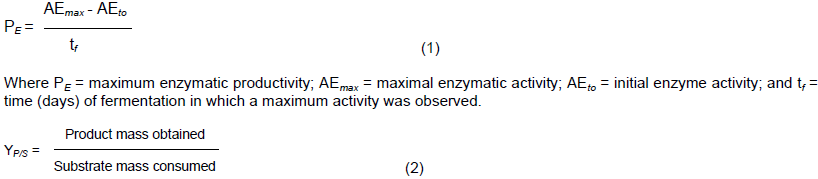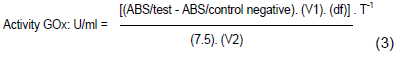ABSTRACT
Glucose oxidase (GOx) has several industrial applications. It is believed that there are several species of fungi that have the ability to produce this enzyme, most of which are unexplored. This work aimed to investigate the production of glucose oxidase (EC 1.1.3.4) by fungi isolated from soil samples of the Amazonian forest. Filamentous fungi were isolated from soil samples from the Adolpho Ducke Forest Reserve, located in Manaus, Amazonas. Strains were subjected to submerged bioprocessing to select for the best GOx producers. Those selected for the production of isolated enzymes were subjected to kinetic tests that evaluated production of the enzyme and consumption of the biomass substrate by the isolates. In addition, experiments to evaluate the optimal carbon, nitrogen and phosphorus sources as well as the influence of the bioprocess factors were carried out. Finally, GOx production was investigated in a semi-continuous system for 7 days. The most frequent isolates isolated from soil samples belonged to the genera Aspergillus, Penicillium and Trichoderma. Aspergillus niger LMM01 was the best GOx producer. Glucose, peptone and KH2PO4 were demonstrated to be the optimal carbon, nitrogen and phosphorus sources, respectively. Multivariate experiments demonstrated that the parameters with the greatest effect on GOx production were pH and agitation. Stable expression results for GOx (7.74 U/ml) were obtained over 7 days in a semi-continuous process. In this context, the new Amazonian source of this enzyme (A. niger LMM01), and enzyme production in a semi-continuous process, demonstrates the importance of the present work.
Key words: Amazon, fungi, production, glucose oxidase.
Glucose oxidase (GOx, β-D-glucose:oxygen 1-oxidoreductase, EC 1.1.3.4) catalyses the oxidation of glucose using molecular oxygen (Bankar et al., 2009b). Since the early 1950s, glucose oxidase has been widely n used in the manufacture of powdered eggs, paper test strips for diabetic patients, and more recently, biosensors (Hwa, 2015; Choi et al., 2015; Wong et al., 2008). Glucose oxidase is produced by fungi as a defence mechanism and has antimicrobial effects (Ferri et al., 2011). This enzyme might also be a good alternative to traditional, chemical and physical treatments used in food preservation (Aber et al., 2016; Amiri et al., 2016; Röcker et al., 2016), but is not highly produced in Brazil.
The most common microbial sources of GOx include members of the Aspergillus and Penicillium genera, although the majority of commercial preparations are made using A. niger species (Qiu et al., 2016; Khan et al., 2016). Extensive growth with the use of GOx in the field of nanotechnology for constructing glucose-sensing biosensors has reduced its importance in the previously mentioned industrial applications (Aber et al., 2016; Sapountzi et al., 2017; Fapyane et al., 2016).
The Amazon rainforest has great potential for bioprospecting useful substances from fungi (Celestino et al., 2014, Pereira et al., 2016). The roles of fungi in soil are complex and critical to the functionality of the biome. Fungi can act in nutrient cycling and develop symbiotic or pathogenic associations with plants and animals in addition to interacting with other microorganisms. The magnitude of the many functions performed by fungi can be better understood by considering the diversity of this group of organisms. Nevertheless, there are no published studies that have screened for GOx producers from the Amazon rainforest (Mendes et al., 2015; Santos et al., 2015; Salony et al., 2016).
The present work describes a GOx produced by A. niger LMM01, representing a new source of GOx. In addition, the isolation of amazon fungi, screening of GOx producers, the optimal bioprocessing conditions and potential use of a semi-continuous system were examined. Academic centres, industry and the general population will likely benefit from the results of this study.
Fungi isolation
Soil samples were collected in June from the florestal reserve Adolpho Ducke (02Ëš55’-03Ëš01’S, 59Ëš53’-59Ëš59’W) Manaus-AM Brazil. Approximately 1 g of superficial soil (2 cm deep) was subjected to successive dilutions (1×10−1-1×10−5) and 100 μL aliquots were plated onto Potato Dextrose Agar (PDA) plates containing chloramphenicol (250 mg/L). The plates were incubated at 30°C and monitored daily for 14 days to isolate filamentous fungi. The isolated colonies were purified and plated on potato dextrose agar (PDA) and incubated at 36°C, during seven days (Gomes et al., 2005). These pure cultures were stored in a refrigerator at 4°C during storage and after the procedure.
Fungi identification
The isolates were transferred to two tubes, one of which was stored at 4°C while the second was used in subsequent experiments. The genera of the isolates were investigated by micromorphological evaluations (Bridge and Spooner, 2001) and the species were identified by sequencing ITS1-5.8S-ITS2 DNA.
DNA was extracted from the samples (200 μL of fungal biomass) using a QIAamp Blood and Tissue kit (Qiagen, Hilden, Germany) following the manufacturer's instructions. DNA was quantified by absorbance at 260 nm using a Genequant Spectrophotometer (Eppendorf, Hamburg, Germany). PCR reactions had a final volume of 50 μL and contained PCR buffer (10 mM Tris-HCl, pH 8.3, 50 mM KCl), 1.5 mM MgCl2, 0.5 µM of the primers ITS-1 (5'-TCCGTAGGTGAACCTGCGG -3') and ITS-4 (5'TCCTCCGCTTATTGATATGC-3'), 200 µM dNTPs, 1.5 U of ampli-tagged DNA polymerase and 100 ng of fungal DNA.
PCR was performed as described by White et al. (1990) using the following PCR amplification conditions in thermocycler (Mastercycler nexus X2, Eppendorf, Brazil): Initial denaturation at 94°C for 5 min, followed by 35 cycles of DNA denaturation at 94°C for 1 min, annealing at 55°C for 1 min and an extension at 72°C for 2 min, with a final extension at 72°C for 10 min. Two positive controls and a negative control were included in the assay. PCR products were visualized by electrophoresis on a 1.5% agarose gel that was stained with SYBR® Green (SYBR Safe DNA Gel Stain, Invitrogen, Carlsbad, USA). A 100-bp DNA Ladder (SM0331, MBI Fermentas, St. Leon-Rot, Germany) was used the DNA size marker.
The amplification products were purified using a polyethylene glycol (PEG) solution (10 g of polyethylene glycol 800, 7.3 g NaCl in 35 ml water) as described previously (Vingataramin and Frost, 2015). Similar volumes of PEG and amplicon solutions were transferred to a microfuge tube (1.5 ml), homogenized and incubated (37°C for 15 min). Then, the mixture was centrifuged (15 min at 6,000 g), supernatant was discarded, and precipitate was washed twice with 70% ethanol and resuspended in water.
The sequencing reaction was performed using a BigDye® kit (Applied Biosystem). Sequencing was carried out in a Seq 3130 Genetic Analyser (Applied Biosystem). The sequences obtained were compared with those in Genbank (www.ncbi.nlm.nih.gov).
Screening of glucose oxidase producers
Fungal strains were incubated in 125-ml flasks containing 50 ml of medium (5 g/L KH2PO4, 0.1 g/L CaCl2, 0.5 g/L MgSO4·7H2O, 0.01 g/L FeSO4·7H2O, 0.01 g/L ZnSO4·7H2O, 0.03 g/L MnSO4·H2O, 5 g/L peptone and 80 g/L glucose, pH 6.0). The culture medium was inoculated with a suspension of spores (1×105 spores/ml). The bioprocess was incubated at 26°C for 96 h with orbital agitation (100 rpm). After incubation, the enzymatic activities were determined.
Kinetics of GOx production of selected isolates
The kinetics of GOx production was determined for microorganisms that excelled at GOx production. The bioprocess conditions were similar to those described in the Screening of Glucose oxidase producers section. Soluble solids (g/L), biomass (g/L) and GOx production (U/ml) were quantified every 24 for 120 h. In addition, GOx productivity (Equation 1) and yield YP/S (Equation 2) were determined:
Sources of carbon, nitrogen and phosphorus
To select the optimal carbon, nitrogen and phosphorus sources, several sources were investigated using a univariate strategy. The main bioprocess conditions were similar to those used in the Screening of Glucose oxidase producers section. The carbon sources (80 g/L) tested included xylose, maltose, glucose, sucrose, galactose and lactose. The nitrogen sources (5 g/L) tested included yeast extract, malt extract, peptone, soy peptone and KNO3. The phosphorus sources (5 g/L) tested was Na3PO4, KH2PO4 and P4O10.
Evaluation of the influence of the assayed factors in the bioprocess through factorial design
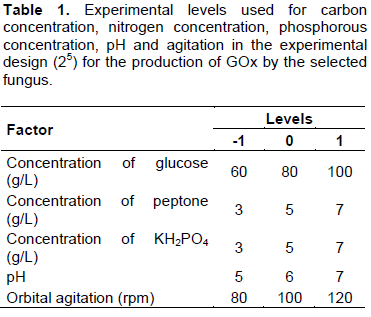
Using a multivariate experiment, the influence of the factors were evaluated: [Carbon Concentration] [Nitrogen Concentration] [Concentration of Phosphorus] [shake] and [pH] in the production of GOx. The bioprocess conditions were similar to those described in the Screening of Glucose oxidase producers section.
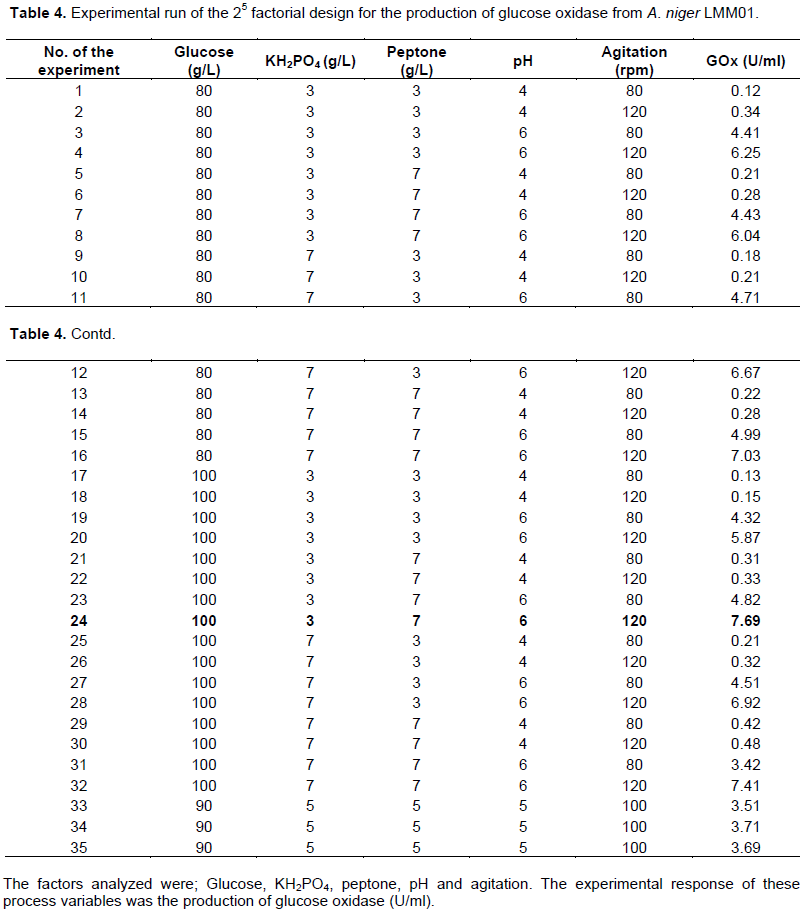
The influence of these variables was studied according to a complete 25 experimental design to identify the factors with statistically significant influence. Thirty-two experiments were carried out, with three central points. A mathematical model was determined and used response surface methodology and central points to calculate the experimental error. The levels of the factors studied are shown in Table 1.
Production in a semi-continuous bioprocess
Cultures were scaled in 2-L flasks containing 0.5 L of medium. The bioprocess was maintained at 27ºC and 120 rpm (agitation speed). The pH of the culture was kept at 6.0. The cultures were grown under ambient light, and the conditions were identical for all growth phases. For semi-continuous operation, after 72 h of initial growth, another growth phase was initiated by following the identical cultivation parameters by removing the fermented medium and replacing it with new medium for 24 h for each assay.
Determination of glucose oxidase
For the determination of GOx a modified method using O-dianisidine, was used (Haq et al., 2014). The fermented broth was centrifuged at 10,000 rpm for 30 min to separate the biomass. For the determination of GOx, a reaction mixture consisting of 0.1 ml of 0.01 M D-glucose, 0.75 ml of 0.02% o-dianisidine in phosphate buffer pH 7.0, 25 µg of horseradish peroxidase (EC 1.11.1. 7) (~40 units/mg solid, Sigma, Germany) and 0.1 ml of enzyme solution (GOx) was used. The mixture was incubated for 10 min at 35°C. The reaction was stopped by adding 0.5 ml of 4 N HCl and quantified at 500 nm. One unit (U) of GOx activity was defined as the amount of enzyme required to oxidize 1 mmol of glucose per minute under the assay conditions. Equation 3 was used to calculate the maximum enzymatic productivity.
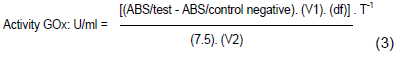
Where, V1 = assay volume (in ml); (df) = dilution factor of the broth; 7.5 = millimolar extinction coefficient for oxidized o-dianisidine at 500 nm; V2 = volume (in ml) of broth containing enzyme used; and T = time duration of the reaction. One unit will oxidize 1.0 mmol of β-D-glucose to gluconolactone and δ-H2O2 per minute at pH 7.0 at 35°C.
Biomass determination
The mycelia were separated from the culture medium by filtration using Whatman No. 1 filter paper and were dried at 105°C for 24 h. The growth of the fungi was quantified in terms of the dry mycelial weight per millilitre. To this end, the mycelia obtained after filtration were transferred to an oven at 105°C for 24 h and then weighed to determine the dry weight.
Sugar determination
The concentration of reducing sugars (RS) was determined by the dinitrosalicylic acid method 3.5 - DNS (Miller, 1959). Samples were centrifuged at 3,500 rpm for 5 min, diluted with H2O and transferred to test tubes containing 1 ml of DNS reagent solution. After incubation in a boiling water bath for 5 min and subsequent cooling with running water, the absorbance of each sample was measured at 540 nm with a digital FEMTO 700S spectrophotometer. The blank reaction used for calibrating the apparatus was obtained using only 1 ml of distilled water. The observed absorbance was correlated to the concentration of reducing sugar using a standard glucose curve (Ahmed et al., 2016). As an alternative, in the semi-continuous process, the Brix was measured in an ATAGO Pocket PAL-1 refractometer (Tokyo, Japan) at 25°C. The refractometer was calibrated using a sucrose solution as a standard, such that 1 Brix was equivalent to 1 g of sucrose per 100 ml of solution.
Isolation and identification
Soil samples were diluted and transferred to culture medium to isolate the fungal strains. Then, identification techniques (determination of micromorphologies and ITS1-5.8S-ITS2 sequencing) were carried out. The total of the 100 isolates primarily belonged to the genera Aspergillus, Penicillium and Trichoderma (Table 2).
GOx screening
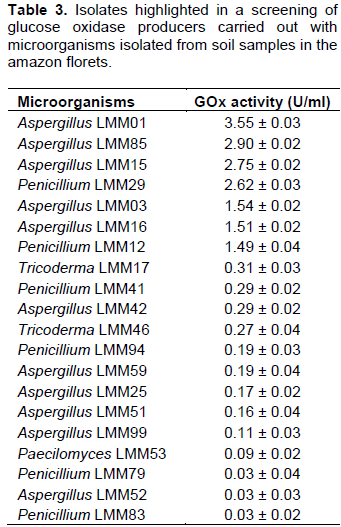
Fungal strains (n=100) were subjected to submerged bioprocess (120 h). The GOx activities (U/ml) of the 20 best producers are shown in the Table 3.
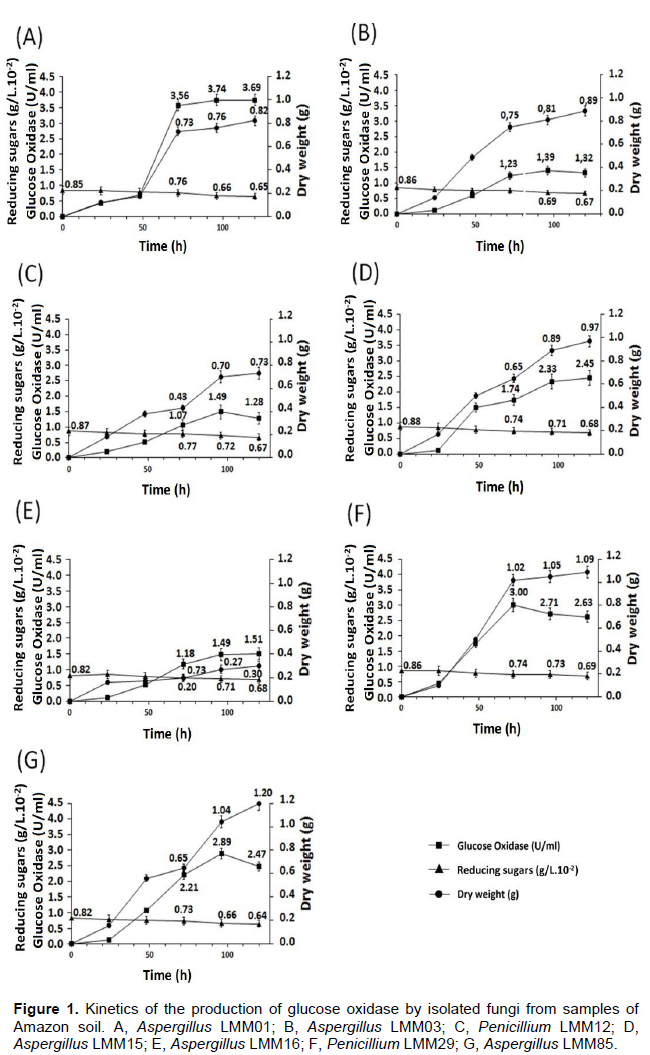
Kinetics of GOx production
GOx (U/ml) (P), fungal biomass (g/L) (X) and reducing sugars (S) were quantified to find the best GOx producer (Figure 1). A. niger LMM01 presented highest productivity (0.093 U/dL) and YP/S (1.87).
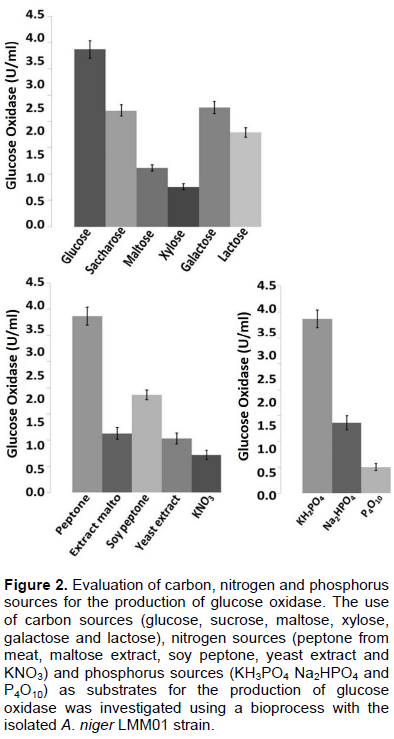
Influence of nutrients and bioprocess conditions
Experiments were carried out to select the optimal carbon, nitrogen and phosphorus sources for GOx production (A. niger LMM01). Glucose, peptone and KH2PO4 were demonstrated to be the optimal carbon, nitrogen and phosphorus sources, respectively (Figure 2).
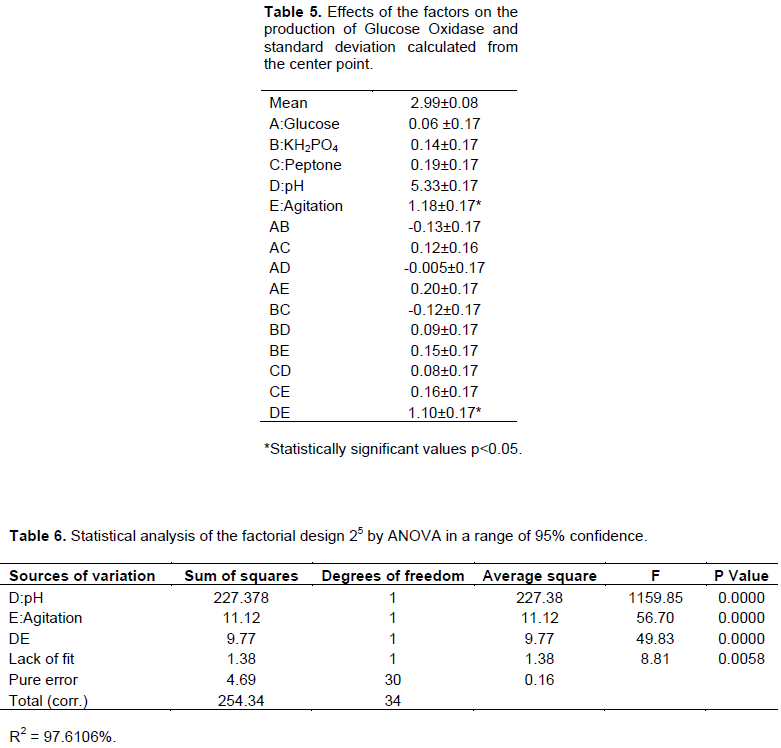
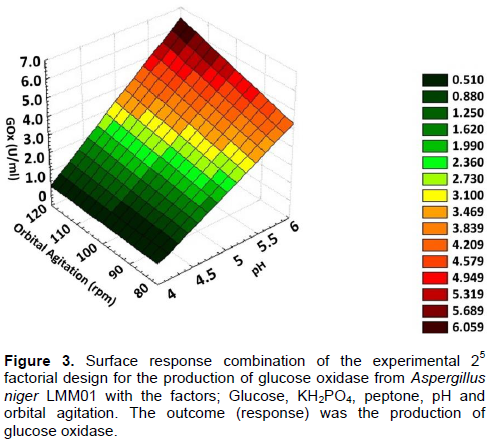
In addition, 25 multifactorial experiments investigated the influence of the following factors on GOx production: [glucose], [peptone], [KH3PO4], orbital agitation (rpm) and pH. The GOx production levels ranged from 0.12 to 7.7 U/ml (Table 4). The effect of the tested factors and their interactions are shown in Table 5. The standard error (s) was calculated and statistical analyses (t-test) were carried out (Table 6). In the experimental conditions, the factors that were considered to be significant for GOx production (95% confidence) were pH (D), agitation (E) and their interaction DE. A mathematical model (Equation 4) was developed according to this data:
GOx (U/ml) = 0.525786 - 0.096875*pH - 0.108656*Agitation + 0.0.27625*pH*Agitation 4
Analysis of variance (ANOVA) was performed (Table 6) to validate the model Equation 4. This mathematical model showed a significant regression and no lack of fit. A graphic of the surface response (Figure 3) was generated to represent the results contained in Equation 4. The response surface showed the highest GOx level at pH 6.0 and an orbital agitation of 120 rpm.
GOx production in a semi-continuous process
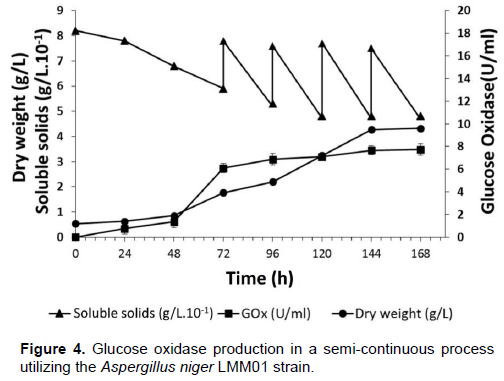
GOx (U/ml) (P), fungi biomass (g/L) (X) and reducing sugars (S) were quantified during the 7-day experiment (Figure 4). Stable results for P (7.74 U/ml), X (9.60 g/L) and S (4.8 g/L) were obtained from the process every 24 h over the course of 7 days.
The present study demonstrated the potential of GOx production by fungal isolates obtained from soil samples from the Amazon region. Specifically, a new isolate, A. niger LMM01, produced high levels of GOx (7.74 U/ml) (Figure 4). In addition, continuous production of this enzyme in a semi-continuous process with high enzymatic levels by using sucrose (as an alternative to glucose) as the substrate was demonstrated. These data demonstrate, for the first time, the potential of strains from Amazonian soil to produce this economically important enzyme.
In the present study, fungal strains isolated from soil samples belonged to the genera Aspergillus, Penicillium and Trichoderma. These fungi were the more prevalent in soil samples from previous studies (Oliveira et al., 2013; Hamed, 2013; Zhang et al., 2014; Iqbal and Utara, 2015; Mirhendi et al., 2016; Sondhia et al., 2016) and are also Prevalent in other environments such as water (Sessegolo et al., 2011; Oliveira et al., 2016), plants (Pinheiro et al., 2013; Zhou et al., 2016; Corrêa et al., 2014; Russell et al., 2011; Orlandelli et al., 2012), marine organisms (Debbab et al., 2012; Subramani et al., 2013; Hayashi et al., 2016; Cicatiello et al., 2016) and even the glacial arctic environment (Sonjak et al., 2006; Sonjak et al., 2007; Zhang et al., 2016).
A fraction of the fungal isolates (20%) were able to produce GOx. This observation is similar to previous GOx screening studies, which also demonstrated the potential of the genus Aspergillus and Penicillium (Gao et al., 2012; Singh and Verma, 2013; Bhat et al., 2013; Ahmad et al., 2014; Konishi et al., 2013; Farid et al., 2012; Gu et al., 2015) to produce GOx. This study is the first to describe a GOx screen carried out with Amazonian fungi.
In the present work, the kinetics study demonstrated that GOx production occurred in the exponential growth phase and that A. niger LMM01 was the best producer. Other studies that investigated GOx production by Aspergillus niger obtained similar results. Bankar et al. (2009b) obtained a GOx yield of 2.0 U/ml; Fiedurek and Gromada (2000) reported a yield of 2.5 U/ml and Lu et al. (1996) obtained a yield of 6.0 U/ml.
Glucose, peptone and KH2PO4 were optimal nutrient sources for GOx production in A. niger LMM01. Hatzinikolaou and Macris (1995) investigated the effects of different carbon sources on GOx production by A. niger and observed that significant enzyme production was obtained using glucose, sucrose and molasses as substrates. Kona et al. (2001) also demonstrated the potential of sucrose as an alternative carbon source. In addition, Rogalski et al. (1988) and Hatzinikolaou and Macris (1995) demonstrated the importance of peptone supplementation for high GOx production.
In the present work, multivariate experiments demonstrated that the parameters that had the greatest effect on the production of GOx were pH and agitation (Table 5), which were observed in previous studies (Ahmad et al., 2014; Röcker et al., 2016). Culture aeration has a direct relationship with the final production of GOx (Macris, 1995; Bankar et al., 2009a; Khurshid et al., 2011). Liu et al. (2003) also demonstrated the importance of aerobic conditions and high oxygen transference for GOx production.
Finally, this study demonstrated the production of high GOx levels in a semi-continuous process during a 7-day assay. Our results demonstrate the potential of A. niger LMM01 and the present bioprocess conditions for use in a wide range of industries, particularly food and pharmaceutical manufacturers (Bankar et al., 2009b; Ferri et al., 2011; Wong et al., 2008).
In recent years, the importance of GOx has increased. Numerous studies (Talbert et al., 2014; Wong et al., 2008; Choi et al., 2015; Singh and Verma, 2013; Röcker et al., 2016) have shown increase in the traditional uses of this enzyme. In addition, GOx has more recently been used in biosensor applications (Devasenathipathy et al., 2015; Turkmen et al., 2014; Hwa, 2015; Lin et al., 2015) and other nanotechnological applications have been demonstrated (Aber et al., 2016; Liu et al., 2014; Sapountzi et al., 2017). In this context, the new Amazonian source of this enzyme (A. niger LMM01) and production of GOx in a semi-continuous process demonstrates the importance of the present work. In the future, scaled up experiments and the utilization of residues as substrate should be investigated to make the enzyme production more economically attractive and their use more widespread.
The authors have not declared any conflict of interests.
REFERENCES
|
Aber S, Mahmoudikia E, Karimi A, Mahdizadeh F (2016). Immobilization of Glucose Oxidase on Fe3O4 Magnetic Nanoparticles and its Application in the Removal of Acid Yellow 12. Water Air Soil Pollut. 227(3):1-11.
Crossref
|
|
|
|
Ahmad I, Islam ZU, Yu Z, Javed MM (2014). Propagation of Aspergillus niger in Stirred Fermentor for the Production of Glucose Oxidase. J. Pure Appl. Microbiol. 8(2):1735-1742.
|
|
|
|
|
Amiri A, Shahedi M, Kadivar M, Aber S, Mahmoudikia E, Karimi A, Grossmann M (2016). Evaluation of physicochemical properties of gluten modified by Glucose oxidase and Xylanase. J. Cereal Sci.
Crossref
|
|
|
|
|
Bankar SB, Bule MV, Singhal RS, Ananthanarayan L (2009b). Glucose oxidase-an overview. Biotechnol. Adv. 27(4):489-501.
Crossref
|
|
|
|
|
Bankar SB, Bule, MV, Singhal, RS, Ananthanarayan, L (2009a). Optimization of Aspergillus niger fermentation for the production of glucose oxidase. Food Bioprocess Technol. 2(4):344-352.
Crossref
|
|
|
|
|
Bhat SV, Swathi BR, Rosy M, Govindappa, M (2013). Isolation and charecterization of Glucose Oxidase (GOD) from Aspergillus flavus and Penicillium sp. Int. J. Curr. Microbiol. Appl. Sci. 2(6):153-161.
|
|
|
|
|
Bridge P, Spooner B. (2001). Soil fungi: diversity and detection. Plant Soil 232(1-2):147-154.
Crossref
|
|
|
|
|
Celestino JR, Carvalho LE, Lima AM, Ogusku MM, Souza JVB (2014). Bioprospecting of Amazon soil fungi with the potential for pigment production. Process Biochem. 49(4):569-575.
Crossref
|
|
|
|
|
Choi JM, Han SS, Kim HS. (2015). Industrial applications of enzyme biocatalysis: Current status and future aspects. Biotechnol. Adv. 33(7):1443-1454.
Crossref
|
|
|
|
|
Cicatiello P, Gravagnuolo AM, Gnavi G, Varese GC, Giardina P (2016). Marine fungi as source of new hydrophobins. Int. J. Biol. Macromol. 92:1229-1233.
Crossref
|
|
|
|
|
Corrêa RCG, Rhoden SA, Mota TR, Azevedo JL, Pamphile JA, de Souza CGM, Peralta RM. (2014). Endophytic fungi: expanding the arsenal of industrial enzyme producers. J. Ind. Microbiol. Biotechnol. 41(10):1467-1478.
Crossref
|
|
|
|
|
Debbab A, Aly AH, Proksch P (2012). Endophytes and associated marine derived fungiecological and chemical perspectives. Fungal Divers. 57(1):45-83.
Crossref
|
|
|
|
|
Devasenathipathy R, Mani V, Chen SM, Huang ST, Huang TT, Lin CM, Chen BJ (2015). Glucose biosensor based on glucose oxidase immobilized at gold nanoparticles decorated graphene-carbon nanotubes. Enzyme Microb. Technol. 78:40-45.
Crossref
|
|
|
|
|
Fapyane D, Poulsen CH, Ferapontova EE (2016). Bioelectrocatalytic oxidation of glucose by hexose oxidase directly wired to graphite. Electrochem. Commun. 65:1-4.
Crossref
|
|
|
|
|
Farid MA, Ghoneimy EA, El-Khawaga MA, Negm-Eldein A, Awad GEA (2012). Statistical optimization of glucose oxidase production from Aspergillus niger NRC9 under submerged fermentation using response surface methodology. Ann. Microbiol. 63(2):523-531.
Crossref
|
|
|
|
|
Ferri S, Kojima K, Sode K (2011). Review of Glucose Oxidases and Glucose Dehydrogenases. J. Diabetes Sci. Technol. 5(5):1068-1076.
Crossref
|
|
|
|
|
Fiedurek J, Gromada A (2000). Production of catalase and glucose oxidase by Aspergillus niger using unconventional oxygenation of culture. J. Appl. Microbiol. 89(1):85-89.
Crossref
|
|
|
|
|
Gao Z, Li Z, Zhang Y (2012). High-level expression of the Penicillium notatum glucose oxidase gene in Pichia pastoris using codon optimization. Biotechnol. Lett. 34(3):507-514.
Crossref
|
|
|
|
|
Gomes E, Souza SR, Grandi RP, Silva RD (2005). Production of thermostable glucoamylase by newly isolated Aspergillus flavus a 1.1 and Thermomyces lanuginosus a 13.37. Braz. J. Microbiol. 36:75-82.
Crossref
|
|
|
|
|
Gu L, Zhang J, Liu B, Du G, Chen J (2015). High-level extracellular production of glucose oxidase by recombinant Pichia pastoris using a combined strategy. Appl. Biochem. Biotechnol. 175(3):1429-1447.
Crossref
|
|
|
|
|
Hamed SAM (2013). In-vitro studies on wood degradation in soil by soft-rot fungi: Aspergillus niger and Penicillium chrysogenum. Int. Biodeterior. Biodegradation 78:98-102.
Crossref
|
|
|
|
|
Haq IU, Nawaz A, Mukhtar H, Ahmed W (2014). Isolation and Identification of Glucose Oxidase Hyper Producing Strain of Aspergillus niger. Br. Microbiol. Res. J. 4(2):195-205.
Crossref
|
|
|
|
|
Hatzinikolaou DG, Macris BJ (1995). Factors regulating production of glucose oxidase by Aspergillus niger. Enzyme Microb. Technol. 17(6): 530-534.
Crossref
|
|
|
|
|
Hayashi A, Crombie A, Lacey E, Richardson A, Vuong D, Piggott A, Hallegraeff G (2016). Aspergillus Sydowii Marine Fungal Bloom in Australian Coastal Waters, Its Metabolites and Potential Impact on Symbiodinium Dinoflagellates. Mar. Drugs 14(3):59.
Crossref
|
|
|
|
|
Hwa K (2015). Immobilization of Glucose Oxidase on Gold Surface for Applications in Implantable Biosensors. J. Med. Bioeng. 4(4):297-301.
Crossref
|
|
|
|
|
Iqbal J, Utara U (2015). Isolation of Aspergillus niger Strains from Soil and their Screening and Optimization for Enhanced Citric Acid Production using Cane Molasses as Carbon Source. J. Appl. Environ. Biol. Sci. 5(4):128-137.
|
|
|
|
|
Khan I, Qayyum S, Ahmed S, Niaz Z, Fatima N, Chi ZM (2016). Molecular cloning and sequence analysis of a PVGOX gene encoding glucose oxidase in Penicillium viticola F1 strain and it's expression quantitation. Gene 592(2):291-302.
Crossref
|
|
|
|
|
Khurshid S, Kashmiri MA, Quershi Z, Ahmad W (2011). Optimization of glucose oxidase production by Aspergillus niger. Afr. J. Biotechnol. 10(9):1674-1678.
|
|
|
|
|
Kona R P, Qureshi N, Pai JS (2001). Production of glucose oxidase using Aspergillus niger and corn steep liquor. Bioresour. Technol. 78(2):123-126.
Crossref
|
|
|
|
|
Konishi T, Aoshima T, Mizuhashi F, Choi SSH, Roberts A (2013). Safety evaluation of glucose oxidase from Penicillium chrysogenum. Regul. Toxicol. Pharmacol. 66(1):13-23.
Crossref
|
|
|
|
|
Lin Y, Hu L, Yin L, Guo L (2015). Electrochemical glucose biosensor with improved performance based on the use of glucose oxidase and Prussian Blue incorporated into a thin film of self-polymerized dopamine. Sens. Actuators B Chem. 210:513-518.
Crossref
|
|
|
|
|
Liu D, Yang J, Wang H, Wang Z, Huang X, Wang Z, Chen X (2014). Glucose Oxidase-Catalyzed Growth of Gold Nanoparticles Enables Quantitative Detection of Attomolar Cancer Biomarkers. Anal. Chem. 86(12):5800-5806.
Crossref
|
|
|
|
|
Liu JZ, Weng LP, Zhang QL, Xu H, Ji LN (2003). Optimization of glucose oxidase production by Aspergillus niger in a benchtop bioreactor using response surface methodology. World J. Microbiol. Biotechnol. 19(3):317-323.
Crossref
|
|
|
|
|
Lu T, Peng X, Yang H, Ji L (1996). The production of glucose oxidase using the waste myceliums of Aspergillus niger and the effects of metal ions on the activity of glucose oxidase. Enzyme Microb. Technol. 229(96):339-342.
|
|
|
|
|
Mendes MMGS, Pereira SA, Oliveira RL, Silva LAO, Albuquerque PM (2015). Screening of Amazon fungi for the production of hydrolytic enzymes. Afr. J. Microbiol. Res. 9(10):741-748.
Crossref
|
|
|
|
|
Miller GL (1959). Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 31(3):426-428.
Crossref
|
|
|
|
|
Mirhendi H, Zarei F, Motamedi M, Nouripour-Sisakht S (2016). Aspergillus tubingensis and Aspergillus niger as the dominant black Aspergillus, use of simple PCR-RFLP for preliminary differentiation. J. Mycol. Med. 26(1):9-16.
Crossref
|
|
|
|
|
Oliveira LG, Cavalcanti MAQ, Fernandes MJS, Lima DMM (2013). Diversity of filamentous fungi isolated from the soil in the semiarid area, Pernambuco, Brazil. J. Arid Environ. 95:49-54.
Crossref
|
|
|
|
|
Oliveira MMC, Marinho BM, Benassi, VM (2016). XII Seminário Brasileiro de Tecnologia Enzimática ENZITEC 2016:2014–2017.
|
|
|
|
|
Orlandelli RC, Alberto RN, Almeida TT, Azevedo JL, Pamphile JA (2012). In vitro Antibacterial Activity of Crude Extracts Produced by Endophytic Fungi Isolated from Piper hispidum Sw. J. Appl. Pharm. Sci. 2 (10):137-141.
Crossref
|
|
|
|
|
Pereira SA, Oliveira RL, Duvoisin Jr S, Silva LADO, Albuquerque PM (2016). The use of Amazon fungus (Trametes sp.) in the production of cellulase and xylanase. Afr. J. Biotechnol. 15(20):843-853.
Crossref
|
|
|
|
|
Pinheiro EAA, Carvalho JM, Dos Santos DCP, Feitosa AO, Marinho PSB, Guilhon G MSP, Marinho AMR (2013). Chemical constituents of Aspergillus sp EJC08 isolated as endophyte from Bauhinia guianensis and their antimicrobial activity. An. Acad. Bras. Cienc. 85(4):1247-1252.
Crossref
|
|
|
|
|
Qiu Z, Guo Y, Bao X, Hao J, Sun G, Peng B, Bi W (2016). Expression of Aspergillus niger glucose oxidase in yeast Pichia pastoris SMD1168. Biotechnol. Biotechnol. Equip. 30(5):998-1005.
Crossref
|
|
|
|
|
Röcker J, Schmitt M, Pasch L, Ebert K, Grossmann M (2016). The use of glucose oxidase and catalase for the enzymatic reduction of the potential ethanol content in wine. Food Chem. 210:660-670.
Crossref
|
|
|
|
|
Rogalski J, Fiedurek J, Szczordrak J, Kapusta K, Leonowicz A (1988). Optimization of glucose oxidase synthesis in submerged cultures of Aspergillus niger G-13 mutant. Enzyme Microb. Technol. 10(8):508-511.
Crossref
|
|
|
|
|
Russell JR, Huang J, Anand P, Kucera K, Sandoval AG, Dantzler KW, Strobel SA (2011). Biodegradation of polyester polyurethane by endophytic fungi. Appl. Environ. Microbiol. 77(17):6076-6084.
Crossref
|
|
|
|
|
Santos YVS, Freire DA, Pinheiro S, Fontão L, Souza JVB, Cavallazzi JRP (2015). Production of laccase from a white rot fungi isolated from the Amazon forest for oxidation of Remazol Brilliant Blue-R. Sci. Res. Essays 10(4):132-136.
Crossref
|
|
|
|
|
Sapountzi E, Braiek M, Vocanson F, Chateaux JF, Jaffrezic-Renault N, Lagarde F (2017). Gold nanoparticles assembly on electrospun poly(vinyl alcohol)/poly(ethyleneimine)/glucose oxidase nanofibers for ultrasensitive electrochemical glucose biosensing. Sens. Actuators B Chem. 238:392-401.
Crossref
|
|
|
|
|
Sessegolo T, Tochetto C, Zanette RA, Schafer A, Alves SH, Monteiro SG, Santurio J M (2011). Fungal microbiota in drinking water and domestic sewage. Semin: Ciênc. Agrár. 32(1):301-306.
Crossref
|
|
|
|
|
Singh J, Verma N (2013). Glucose oxidase from Aspergillus niger : Production, characterization and immobilization for glucose oxidation. Adv. Appl. Sci. Res. 4(3):250-257.
|
|
|
|
|
Sondhia S, Rajput S, Varma RK, Kumar A (2016). Biodegradation of the herbicide penoxsulam (triazolopyrimidine sulphonamide) by fungal strains of Aspergillus in soil. Appl. Soil Ecol. 105:196-206.
Crossref
|
|
|
|
|
Sonjak S, Frisvad JC, Gunde-Cimerman N (2006). Penicillium mycobiota in Arctic subglacial ice. Microb. Ecol. 52(2):207-216.
Crossref
|
|
|
|
|
Sonjak S, UršiÄ V, Frisvad JC, Gunde-Cimerman N (2007). Penicillium svalbardense, a new species from Arctic glacial ice. Antonie van Leeuwenhoek 92(1):43-51.
Crossref
|
|
|
|
|
Subramani R, Kumar R, Prasad P, Aalbersberg W (2013). Cytotoxic and antibacterial substances against multi-drug resistant pathogens from marine sponge symbiont: Citrinin, a secondary metabolite of Penicillium sp. Asian Pac. J. Trop. Biomed. 3(4):291-296.
Crossref
|
|
|
|
|
Talbert JN, He F, Seto K, Nugen SR, Goddard JM (2014). Modification of glucose oxidase for the development of biocatalytic solvent inks. Enzyme Microb. Technol. 55:21-25.
Crossref
|
|
|
|
|
Turkmen E, Bas SZ, Gulce H, Yildiz S (2014). Glucose biosensor based on immobilization of glucose oxidase in electropolymerized poly(o-phenylenediamine) film on platinum nanoparticles-polyvinylferrocenium modified electrode. Electrochim. Acta 123:93-102.
Crossref
|
|
|
|
|
Vingataramin L, Frost EH (2015). A single protocol for extraction of gDNA from bacteria and yeast. Biotechniques 58(3):120-125.
Crossref
|
|
|
|
|
White TJ, Bruns T, Lee S, Taylor J (1990). Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR protocols: a guide to methods and applications, Academic Press. 18(1):315-322.
|
|
|
|
|
Wong CM, Wong KH, Chen XD (2008). Glucose oxidase: natural occurrence, function, properties and industrial applications. Appl. Microbiol. Biotechnol. 78(6):927-938.
Crossref
|
|
|
|
|
Zhang T, Wang NF, Zhang YQ, Liu HY, Yu LY (2016). Diversity and Distribution of Aquatic Fungal Communities in the Ny-Ålesund Region, Svalbard (High Arctic). Microb. Ecol. 71(3):543-554.
Crossref
|
|
|
|
|
Zhang Z, Liu JL, Lan JY, Duan CJ, Ma QS, Feng JX (2014). Predominance of Trichoderma and Penicillium in cellulolytic aerobic filamentous fungi from subtropical and tropical forests in China, and their use in finding highly efficient β-glucosidase. Biotechnol. Biofuels 7(1):107.
Crossref
|
|
|
|
|
Zhou M, Zhou K, He P, Wang KM, Zhu RZ, Wang YD, Hu QF (2016). Antiviral and Cytotoxic Isocoumarin Derivatives from an Endophytic Fungus Aspergillus oryzae. Planta Med. 82(5):414-417.
Crossref
|
|