ABSTRACT
Clostridium difficile infection (CDI) is a significant health problem worldwide. Control and prevention strategies of C. difficile horizontal transmission require assays with fast detection with high specificity and sensitivity. Conventional diagnostic methods are time consuming and costly for clinical field settings. This study aims to develop gold nanoparticles (AuNPs)-based assay for direct qualitative detection of the nucleic acid of C. difficile and its toxins. A colloidal solution of AuNPs with a diameter of 13±1 nm was prepared and characterized. The qualitative colorimetric AuNPs assay was developed for restricted genomic C. difficile DNA detection, and results were confirmed by PCR. One hundred and five positive C. difficile isolates were collected from patients with diarrheal diseases and tested using AuNPs based-assay. Ninety-six samples (91.4%) were detected positive using AuNPs based assay, as indicated by the color change from red to blue within 1 min. All ninety-six positive samples were positive for toxin B. In conclusion, nano-gold assay prototype was developed for direct and inexpensive detection of C. difficile. The developed prototypes are simple, sensitive, rapid and can substitute PCR-based detection. The developed assay may show potential in the clinical diagnosis of C. difficile, especially in developing countries as it is less costly as compared to the commercially available assays.
Key words: Gold nanoparticles, Clostridium difficile, colorimetric assay, polymerase chain reaction (PCR).
Abbreviation:
CDI, Clostridium difficile Infection; AuNPs, gold nanoparticles; EIAs, enzyme immunoassays; GDH, glutamate dehydrogenase enzyme; LSPR, localized surface plasmon resonance; SEM, scanning electron microscope; RT-PCR, reverse transcription-polymerase chain reaction.
Clostridium difficile is a Gram-positive, strictly anaerobic, spore-forming bacterium (Chankhamhaengdecha et al., 2013), first recognized in the late 1970s (Burnham and Carroll, 2013). It is documented as the causative agent of
a broad spectrum of intestinal diseases ranging from mild self-limiting diarrhea to more serious and potentially life threatening manifestations such as Pseudomembranous colitis and is responsible for most cases of antibiotic-associated diarrhea (Carter et al., 2007; Goncalves and Decre, 2004). Pathogenicity of C. difficile is linked to two major toxins produced by the bacteria: toxins A (enterotoxin) and B (cytotoxin). Toxins A and B are encoded by the genes tcdA and tcdB, respectively, and are located in the 19.6-kb pathogenicity locus (PaLoc) of the C. difficile chromosome (Persson et al., 2011). Some strains of C. difficile also secrete binary toxin. CdtA and cdtB genes that are located outside the PaLoc (Goncalves and Decre, 2004) encode these binary toxins.
Rapid and reliable identification of toxigenic C. difficile is essential for appropriate patient management and implementation of timely infection control measures due to the rapidly increasing infection rate of C. difficile in health care facilities. Current laboratory diagnosis remains challenging with many limitations, as rapid test procedures relying on enzyme immunoassays (EIAs) show limited sensitivity. On the other hand, the gold standard toxigenic culture and cytotoxicity assays, which are considered as reference standard, are time-consuming (Dalpke et al., 2013). A two-step algorithms consisting of sensitive detection of glutamate dehydrogenase enzyme (GDH) followed by a confirmatory test using PCR have been proposed by Ticehurst et al. (2006) to increase sensitivity and specificity of the detection.
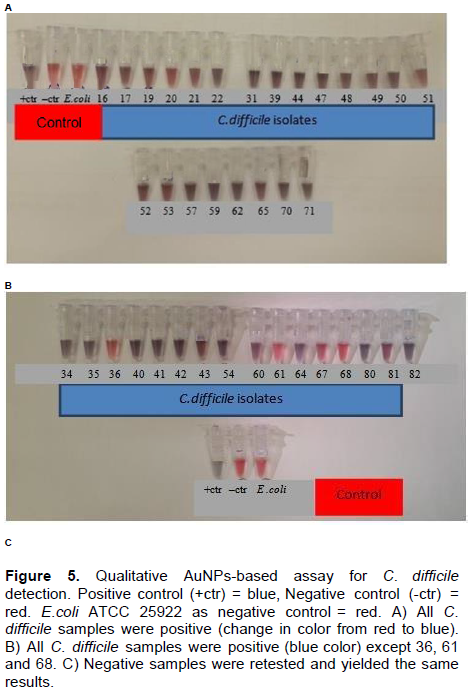
Gold has been used as a vital material in nanotechnology and incorporated in diagnostic procedures. Distinctive size-dependent optical properties of AuNPs, their inertness and strength make them one of the most robust materials utilized in nano diagnostics technology (Syed, 2014). They are spherical in shape with a typical diameter of approximately 2 to 100 nm that exhibit a unique phenomenon known as “localized surface plasmon resonance” or LSPR, which is responsible for their intense red color. Upon aggregation, AuNPs change color and that is easily detected visually without the aid of any instrumentation. Addition of salt (NaCl) during hybridization shields the surface charge on the AuNPs, which are typically negatively charged due to reduced citrate ions on their surfaces, leading to aggregation of AuNPs and thus change in color from red to blue (Hussain et al., 2013; Syed, 2014). This property is especially useful in colorimetric detection based assay, which is evaluated in this study (Azzazy et al., 2006; Jennings and Strouse, 2007; West and Halas, 2003). The principle of colorimetric AuNPs-based assay in microbial identification is illustrated in Figure 1.
To the best of the authors’ knowledge, this is the first study aimed to develop unmodified AuNPs - based assay for direct qualitative detection of the nucleic acid of C. difficile and its toxins. Additionally, it evaluated AuNPs assay sensitivity and specificity as compared to RT-PCR (GeneXpert, Cepheid, CA, USA).
Synthesis of AuNPs
Spherical AuNPs were prepared by citrate reduction of gold 111 chloride trihydrate (HAuCl4.3H2O). Synthesis was carried out as previously prescribed by Grabar et al. (1995). Briefly, the reflux system was cleaned by aqua regia and then rinsed with ultrapure water and left to dry. An aqueous solution of 1 Mm HAuCl4.3H2O (Sigma Aldrich) was brought to boil under reflux conditions while stirring. When gold started to boil, 1% of trisodium citrate (Sigma Aldrich) was rapidly added. This resulted in a subsequent change in solution color from yellow - clear - black - purple - deep red. Afterwards, the solution was then refluxed for an additional 15 min and subsequently allowed to cool to room temperature. The colloidal solution was then stored into a clean dark storage glass bottle at 4°C until further use.
Characterization of AuNPs
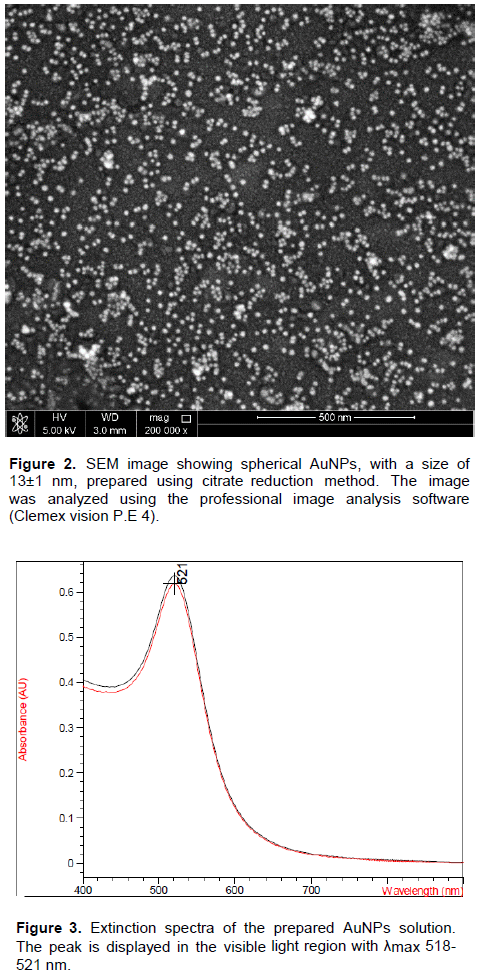
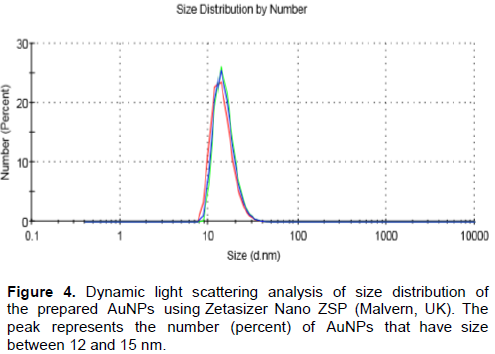
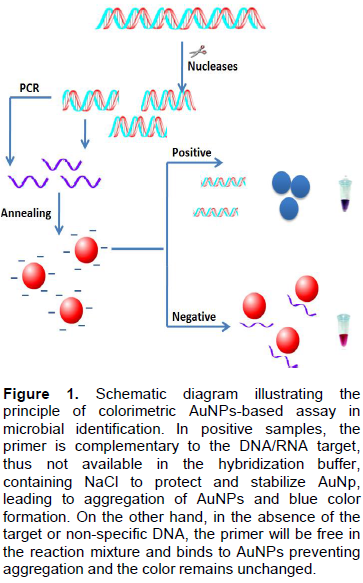
The absorbance of the prepared AuNPs solution was measured by Agilent 8453 UV-visible spectrophotometer at wavelength 400 to 700 nm. The size and distribution was characterized using Zeta sizer (Malvern, Nano ZSP, UK) based on light scattering principle and scanning electron microscope, SEM (FE1 NOVA-NanoSEM, 450, USA). The recommended AuNPs size ranges from 12 to 15 nm with the absorbance of the visible range 400 to 700nm (Grabar et al., 1995).
Collection of bacterial isolates
A total of 105 C. difficile clinical isolates used in this study were, originally provided for routine laboratory diagnosis at Hamad Medical Corporation (HMC) - Al Khor Hospital, Qatar during the period of 2011 to 2012. Cryopreserved C. difficile isolates were revived and sub-cultured onto blood agar enrichment medium, and incubated anaerobically. Samples were transported to Qatar University (Biomedical Research Center) for DNA extraction and testing with gold nanoparticles-based assay. Isolates were confirmed to be C. difficile using RT-polymerase chain reaction, RT-PCR, (GeneXpert, Cepheid, CA, USA). According to RT- PCR results, all C. difficile isolates (105) were positive for toxin B. Twenty nine additional ATCC bacterial isolates other than C. difficile were used to assess specificity of the assay. The Research Ethical Committee of Hamad Medical Corporation, Qatar, approved the study.
DNA extraction
Genomic DNA was extracted from bacterial cultures following manufacturer's instructions of QIAamp DNA Mini kit (Qiagen; Cat. No. 51306).
Restriction of genomic C. diff DNA
The entire extracted genomic C. diff DNA was digested using Bam HI, according to the conditions recommended by the supplier of the enzyme kit (Promega). Briefly, 17.3 µl of the extracted genomic DNA were restricted by addition of 0.5 µl of the enzyme, 2 µl of the buffer, 0.2 µl of the acetylated bovine serum albumin. Then incubated at 37°C for 1 h after that inactivated at 65°C for 20 min.
Colorimetric AuNPs assay for detection of C. difficile DNA: Development and optimization
The colorimetric qualitative AuNPs assay for C. difficile was optimized through adjustment of the assay parameters such as annealing temperature, salt concentration and targeting oligonucleotide sequences. Different concentrations of NaCl and primer concentrations were tested to determine the optimum concentrations for performing the assay (data not shown). Hybridization buffer was prepared using 0.50 M NaCl and 10 μM primer. Different volumes of the AuNPs were tested, and 25 μL of the prepared AuNPs (12 to 15 nm) was selected for use in the final assay. Forward CD-R 5’- CCC TGC ACC CTT AAT AAC TTG ACC- 3’ (Integrated DNA Technologies, Inc. Belgium) primer was used in the assay due to its high specificity to all C. difficile. The assay was performed as follows: 22 μL of the extracted DNA (1.7ng/μl), were placed in a sterile PCR tube. Then, 13 μL of the hybridization buffer were added and mixed well (final concentration of the primer and NaCl after addition of AuNPs were 0.9 μM and 0.04 M, respectively) to obtain a final volume of 35 μl per PCR tube. The mixture was then heated at 95°C for 30 s, and annealed at 50°C for 30 s and then cooled to room temperature for 10 min. 25 μl of colloidal AuNPs were then added to the mixture, and the color was observed within 1 min (Shawky et al., 2010).
Colorimetric AuNPs assay for detection of C. difficile toxin B
All C. difficile positive samples were further tested for toxin B following similar methods for detection of C. diff DNA by AuNPs but using specific toxin B (Ted B) primer (5-CAC GCC TGG AGA ATC TAT ATT TGT AGA AA-3).
Characterization of AuNPs
The SEM image (Figure 2) showed that the synthesized AuNPs prepared in our laboratory were well dispersed and spherical in shape. The extinction spectrum of the prepared AuNPs demonstrated a single peak in the visible region (400 to 700nm) with λmax at 519 to 521 nm (Figure 3). The diameter of AuNPs was found to be 13±1 nm (Figure 4), as characterized by both SEM and Zeta sizer.
C. difficile gold nanoparticles assay prototype
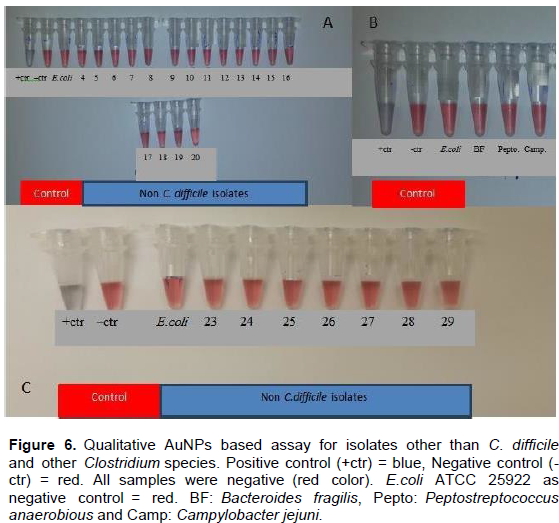
Optimization of the AuNPs-based assay is affected by four main factors namely, concentrations of NaCl, AuNPs size, primer and the assay annealing temperature. In the positive samples, blue color indicates the presence of primers complementary to the C. difficile DNA sequence and this leads to the aggregation of AuNPs. On the other hand, red color indicates that the primer is free in the mixture that leads to stabilization of AuNPs and prevents their aggregation. Any minimal change in color from red to blue or purple is considered as a positive result (Figure 5). Ninety six (96) out of 105 (91.4%) C. diff positive samples showed blue and 9 out of 105 (8.6%) showed a red color. The nine negative samples were retested and yielded same results (Figure 6). Reference strain E. coli ATCC 25922, Bacteroides fragilis, Peptostreptococcus anaerobious and Campylobacter jejuni tested negvative with our assay as expected (Figure 6B). Additionally, all twenty-nine ATCC strains of Clostridium species other than C. diff were also tested negative with AuNPs-based assay (red color) (Figures 6A, B and C) indicating high specificity.Control.
Assay performance assessment
No cross-reactivity was observed between C. difficile and other bacteria tested (Figure 6). The AuNPs-based assay performance was then compared with commercially available RT-PCR (GeneXpert, Cepheid, USA), and yielded 91.4% positivity, all samples were positive with RT-PCR.
Gold nanoparticles assay to detect C. diff toxin B
The assay was then optimized to detect toxin B gene(tcdB) of C. difficile. All ninety-six samples positive with C. difficile prototype detection assay were also positive for toxin B gene (Figure 7). Equivalent result was obtained when using RT-PCR method. C. perfringens ATCC 13124 was used as a negative control in toxin B detection with AuNPs.
AuNPs-based methods have been established for detection of several pathogenic organisms such as Mycobacterium tuberculosis (MTB), Hepatitis C virus (HCV), methicillin resistant Staphylococcus aureus (MRSA) and others, owing to the high sensitivity and specificity of AuNPs- based assays for detection of nucleic acid targets (Shawky et al., 2010; Hussain et al., 2013). AuNPs study carried out by Shawky et al. (2010), showed sensitivity and specificity of 93.3 and 88.9%, respectively, in detecting HCV. Another study by Hussain et al. (2013). indicated 96.6% sensitivity and 98.9% specificity for the detection of M. tuberculosis complex (MTBC) and 94.7% sensitivity and 99.6% specificity for the detection of MTB. These results are consistent with our findings in this study which, demonstrated 91.4% sensitivity and 100% specificity as compared to RT-PCR. PCR assays include amplification steps and has always been considered more sensitive than enzymetic/cloromertic assays. Additionaly, the lower sensitivity of AuNPs- assay (91.4%) could be clarified by the following reasons, 1) possible loss of the targeted genes due to long cryoperservation period, 2) PCR does not cover the same region used to design the primers for PCR.
Different gold nanoparticles approaches have been explored to detect C. difficile and its toxins, such as using an aptamer biosensor with gold nanoparticles synthesized by Bacillus stearothermophilus (Luo et al., 2013). Additionally, Zhu et al. (2015) considered the use of single domain antibody coated gold nanoparticles as enhancer for C. difficile toxin detection by electro chemical impedance immunosensors. This confirms that using AuNPs with different methodologies resulted in excellent performance with high sensitivity and is in concordance with our findings.
This study revealed that AuNPs-based assay could be used to detect C. difficile and C. difficile toxin B from unamplified genomic DNA with detection limit of 35.5 ng DNA. The assay was highly sensitive, specific, rapid, simple and minimized the need for expensive and complex equipment. Cost effectiveness is one of the advantages of synthesis of AuNPs that was explored in this study. The approximate cost of 1 g of gold chloride is 220 US Dollars which is enough to conduct 40,000.00 assays, as compared to ~ 55 USD per test for RT-PCR, (GeneXpert, Cepheid, CA, USA). Additionally, the prepared gold nanoparticles have a long shelf life and can be stored at 4°C for one year or more. Moreover, this method has short turnaround time, only 15 to 20 min, after DNA extraction; consequently, the method can be utilized by many laboratories especially in low-income countries with low resources. This developed assay may improve the management of C. difficile infection by early isolation of infected patient to prevent horizontal transmission in health care facilities, and may lead to a more rational use of antibiotics, as the clinicians will rapidly obtain the clinical microbiology results.
In conclusion, this assay will have a crucial and great impact on clinical diagnosis in low-resources countries in terms of patient’s management and infection control measures. Moreover, the assay can compete with RT-PCR since it has comparable performance results.
The authors have not declared any conflict of interests.
REFERENCES
|
Azzazy HM, Mansour MM, Kazmierczak SC (2006). Nanodiagnostics: a new frontier for clinical laboratory medicine. Clin. Chem. 52(7):1238-1246.
Crossref
|
|
|
|
Burnham CAD, Carroll KC (2013). Diagnosis of clostridium difficile infection: An ongoing conundrum for clinicians and for clinical laboratories. Clin. Microbiol. Rev. 26(3):604-630.
Crossref
|
|
|
|
Carter GB, Lyras D, Allen DL, Mackin KE, Howarth PM, Rood JI (2007). Binary toxin production in Clostridium difficile is regulated by CdtR, a LytTR family response regulatorâ–¿. J. Bacteriol. 189(20):7290-7301.
Crossref
|
|
|
|
Chankhamhaengdecha S, Hadpanus P, Aroonnual, A, Ngamwongsatit, P, Chotiprasitsakul D, Chongtrakool P, Janvilisri T (2013). Evaluation of multiplex PCR with enhanced spore germination for detection of clostridium difficile from stool samples of the hospitalized patients. BioMed Res Int.
Crossref
|
|
|
|
Dalpke AH, Hofko M, Zorn M, Zimmermann S (2013). Evaluation of the fully automated BD MAX C.diff and Xpert C. difficile assays for direct detection of Clostridium difficile in stool specimens. J. Clin. Microbiol. 51(6):1906-1908.
Crossref
|
|
|
|
Goncalves C, Decre D (2004). Prevalence and Characterization of a Binary Toxin (Actin-Specific ADP-Ribosyl transferase) from Clostridium difficile. J. Clin. Microbiol. 42(5):1933-1939.
Crossref
|
|
|
|
Grabar KC, Freeman RG, Hommer MB. Natan MJ (1995). Preparation and characterization of Au colloid monolayers. Anal. Chem. 6:735-43.
Crossref
|
|
|
|
Hussain MM, Samir, TM, Azzazy HME (2013). Unmodified gold nanoparticles for direct and rapid detection of Mycobacterium tuberculosis complex. Clin. Biochem. 46(7-8):633-637.
Crossref
|
|
|
|
Jennings T, Strouse G (2007). Past, present, and future of gold nanoparticles. Adv. Exp. Med. Biol. 620:34-47.
Crossref
|
|
|
|
Luo c, Tang H, Cheng W, Yan L, Zhang, D, Ju, H, Ding S (2013). A sensitive electrochemical DNA biosensor for specific detection of Enterobacteriaceae bacteria by Exonuclease III-assisted signal amplification. Biosens. Bioelectron. 48:132-133.
|
|
|
|
Persson S, Jensen JN., Olsen KEP (2011). Multiplex PCR method for detection of Clostridium difficile tcdA, tcdB, cdtA, and cdtB and internal in-frame deletion of tcdC. J. Clin. Microbiol. 49(12):4299-300.
Crossref
|
|
|
|
Shawky SM, Bald D, Azzazy HME (2010). Direct detection of unamplified hepatitis C virus RNA using unmodified gold nanoparticles. Clin. Biochem. 43:1163-1168.
Crossref
|
|
|
|
Syed MA (2014). Advances in nanodiagnostic techniques for microbial agents. Biosen. Bioelectron. 51:391-400.
Crossref
|
|
|
|
Ticehurst JR, Aird DZ, Dam LM, Borek AP, Hargrove JT, Carroll KC (2006). Effective detection of toxigenic Clostridium difficile by a two-step algorithm including tests for antigen and cytotoxin. J. Clin. Microbiol. 44:1145-1149.
Crossref
|
|
|
|
West JL, Halas NJ (2003). Engineered nanomaterials for biophotonics applications: improving sensing, imaging, and therapeutics. Annu. Rev. Biomed. Eng. 5:285-292.
Crossref
|
|
|
|
Zhu Z, Shi L, Feng H, Zhou HS (2015). Single domain antibody coated gold nanoparticles as enhancer for Colstridium difficile toxin detection by electrochemical impedance immunosensors. Bioelectrochemistry 101:153-158.
Crossref
|