ABSTRACT
This study aimed to assess antimicrobial susceptibility of members of the family Flavobacteriaceae isolated from Nile tilapia (Oreochromis niloticus). Antimicrobial susceptibility of 67 Flavobacteriaceae isolates originating mainly from ponds and Lake Victoria against 19 antimicrobial agents was determined by the broth micro dilution method. Overall, most isolates were susceptible to enrofloxacin (97%; MIC90 2 μg/ml) followed by novobiocin (85%, MIC90, 4 μg/ml) and the aminoglycoside streptomycin (85%; MIC90, 128 μg/ml). Some isolates were also susceptible towards trimethoprim/sulfamethoxazole (77.6%), neomycin and florfenicol both at 62.7%. Susceptibility levels were low for tylosin tartrate (32.8%), clindamycin and sulphathiazole both at (23.9%), ceftiofur (6%), spectinomycin (6%) and tetracyclines/oxtetracyclines (4.5%). In contrast, β-Lactams (amoxicillin, penicillin), gentamycin and erythromycin exhibited very poor activity against Flavobacteriaceae isolates. The extent of antimicrobial susceptibility did not vary significantly among isolates from farmed and wild fish isolates (P > 0.01). The highest Multiple Antimicrobial Resistance (MAR) index was observed in Chryseobacterium indologenes (0.89) and the lowest in Chaetoderma indicum isolates (0.32). Our results indicate that most of Flavobacteriaceae isolates are multidrug resistance, and this may be associated with intrinsic resistance mechanisms to a broad range of antimicrobial agents. However, the need remains to carryout in-depth study to understand better the underlying genetic mechanisms given that the magnitude and trend for susceptibility was comparable between isolates from aquaculture and fisheries. The findings from this study give us insight into appropriate choice of antimicrobial agents for effective treatment of infections caused by these isolates.
Key words: Aquaculture, Fisheries, Intrinsic resistance, minimum inhibitory concentrations, ponds, Lake Victoria.
The genera Flavobacterium and Chryseobacterium belong to the family Flavobacteriaceae and are widely distributed in various environments, including soil, freshwater and saltwater ecosystems (Kumru et al., 2017; McBride, 2014). Several species within the Flavobacteriaceae are regarded as opportunistic pathogens, yet with potential to cause diseases in a wide variety of organisms, including plants, fish and humans (Bernardet and Bowman, 2011; Bernardet and Nakagawa, 2006; Loch and Faisal, 2015). Typically, many of these bacteria cause diseases in fish when water temperatures are relatively high. Excessive organic matter in ponds and high stocking density are among other contributing factors to diseases that consequently lead to economic losses (Loch et al., 2013). In addition, these bacteria have also been associated with nosocomial infections and septicaemia in humans (Loch et al., 2013; Ratnamani, 2013).
Fish farming has grown worldwide as a source of high quality protein and employment to skilled and unskilled workers. In Tanzania, aquaculture is dominated by pond culture of Nile tilapia (Oreochromis niloticus). As aquaculture has developed, a range of bacterial diseases have been encountered (Pathmalal et al., 2018). These diseases are considered as critical limiting factors in this industry, where studies have been conducted and efforts has been given to therapeutic and prophylactic use of antimicrobial agents as control measures of bacterial diseases (Cabello et al., 2006). Many European countries are governed by legislation and regulation on the use of antimicrobial agents in aquaculture (Cabello et al., 2013). However, guidelines for the correct and prudent use of antimicrobial agents in aquaculture have yet to be developed in most of the African countries. There is no list of licensed antimicrobials for use in aquaculture in Tanzania. Previous work has shown that extensive use of antimicrobials in aquaculture leads to resistance across the entire microbial water ecosystem (Schmidt et al., 2000, Watts et al., 2017). In the family Flavobacteriaceae, some antimicrobial resistance has been associated with the presence of plasmids or mutations in specific resistance determinants (Izumi et al., 2004, Madsen et al., 2000). However, other studies have associated antimicrobial resistance in Flavobacteriaceae with intrinsic resistance to a wide range of antimicrobial agents through mechanisms like restricted outer membrane permeability, efflux systems that pump antimicrobials out of the cell and production of antimicrobial-inactivating enzymes such as β-lactamases (Clark et al., 2009; Henríquez-Núñez et al., 2012; Michel et al., 2005).
The occurrence of resistant bacteria to one or more antimicrobials have been reported not only in pathogenic fish bacteria but also among opportunistic bacteria, even in the absence of selective antimicrobial pressure in the aquaculture environment (Gufe et al., 2019; Jensen et al., 2008). In Tanzania, the nutritional benefits of fish consumption have a positive link to increased food security, and the aquaculture sector has the potential to play a significant role in this aspect. However, a sustainable fish production has to be secured, including the development of control measures to reduce the spread of resistant bacteria in the aquaculture environments. As very few data are currently available, the aim of this study was to assess the antimicrobial susceptibility of Flavobacteriaceae isolates from Nile tilapia sampled directly from fish farms as well as natural lacustrine environment in Tanzania. Findings from this study would contribute to our understanding of antimicrobial susceptibility in the Flavobacteriaceae; provide basic knowledge needed in implementation of biosecurity measures in fish farming and give a background to treatment of farmed fish if necessary.
Bacterial isolates from Tanzania
A total of 67 Flavobacteriaceae isolates previously recovered from 40 Nile tilapia farmed in the Morogoro region, and from batch of 21 wild Nile tilapia collected from natural environment in Lake Victoria in Mwanza region. Apparently healthy fish were sampled between November 2015 and May 2016, isolation and identification of the bacteria were performed as previously described in our earlier work (Mwega et al., 2018). Briefly, all isolates were grown on modified Anacker and Ordal agar - AOA (Pilarski et al., 2008). Identification of isolates based on colony morphology and biochemical testing, and was confirmed by 16S rRNA gene sequencing (Darwish et al., 2004). All isolates used in this study were designated with the organism code based on sampling location as shown in Table 3.
Antimicrobial susceptibility tests
Antimicrobial susceptibility to 19 antimicrobials commonly used in veterinary medicine and aquaculture in most developed countries was assessed for all isolates using Trek Sensititre Avian susceptibility plates (Trek Diagnostic Systems, Cleveland, OH) as shown in Table 1. These test plates were 96-well, dry-form that contained twofold serial dilutions of the antimicrobial agents listed in Table 1. Briefly, ten microliters of the bacteria suspension were transferred into a tube containing 11 ml of Sensititre MH broth to give an inoculum of 1×105 CFU/ml. The broth was poured into a sterile seed trough and individual wells inoculated with 50 μl using a multi-channel pipette. Inoculated plates were sealed and incubated aerobically ATCC 25922 and Aeromonas salmonicida NCIMB 1102 were also included in parallel in all testing.
Data management and analysis
In the absence of published resistance breakpoints for Flavobacteriaceae, quantitative interpretation of minimum inhibition concentration (MIC) results was based on CLSI guidelines (CLSI, 2014) with the minor modifications as previously recommended (Akinbowale et al., 2006). Minimum inhibition concentration was recorded as the lowest concentration of antimicrobial that inhibited visible growth. The MIC50 and MIC90 represent the MIC value at which ≥ 50% and ≥ 90% of the isolates were inhibited respectively. A multiple antimicrobial resistance (MAR) index was then determined for each isolate by dividing the number of antimicrobials to which an isolate was resistant with the total number of antimicrobials tested. The MAR index analysis was used to group the different sources from which the bacteria were recovered using the frequency of antimicrobial resistance. Isolates with MAR index < 0.2 were considered as isolates recovered from low-risk sources of contamination while isolates with MAR index > 0.2 were considered recovered from high- risk sources (Samuel et al., 2011; Tambekar et al., 2006).

Antimicrobial susceptibility of Flavobacteriaceae isolates
The in vitro activities of the 19 antimicrobial agents tested against the Flavobacteriaceae isolates are summarized in Table 1, showing both the MIC range and the MIC50 and MIC90 results of all isolates, as well as the number and percentage of susceptible isolates. Figure 1 shows the MIC distribution of the isolates towards streptomycin, enrofloxacin, novobiocin and trimethoprim/ sulfamethoxazole. Among 67 isolates tested, 97% were susceptible to enrofloxacin (MIC50, 0.12 μg/ml, MIC90 2 μg/ml). Novobiocin (MIC90, 4 μg/ml) and streptomycin (MIC90, 128 μg/ml) inhibited 85% of the isolates at the susceptible breakpoints. Moderate susceptibility (60 - 80%) was observed to trimethoprim/ sulfamethoxazole 77.6%, neomycin and florfenicol both at 62.7% of overall isolates. Extent of susceptibility was low (less than 50 - 60%) for tylosin tartrate (32.8%), clindamycin and sulphathiazole both at (23.9%). The susceptibility levels were very low (less than 10%) to ceftiofur (6%), spectinomycin (6%) and tetracyclines/ oxtetracyclines (4.5%). In contrast, 0% overall susceptibility was exihibited for β-Lactams (amoxicillin, penicillin), gentamycin and erythromycin against Flavobacteriaceae isolates at the given susceptible breakpoints (Table 1). MIC values of control strains were within the acceptable range (CLSI, 2014)(Figure 1).
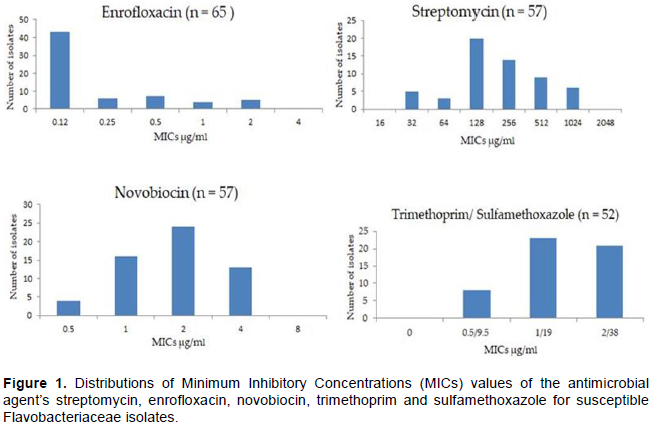
Antimicrobial susceptibility of Flavobacteriaceae isolates according to the sampling groups
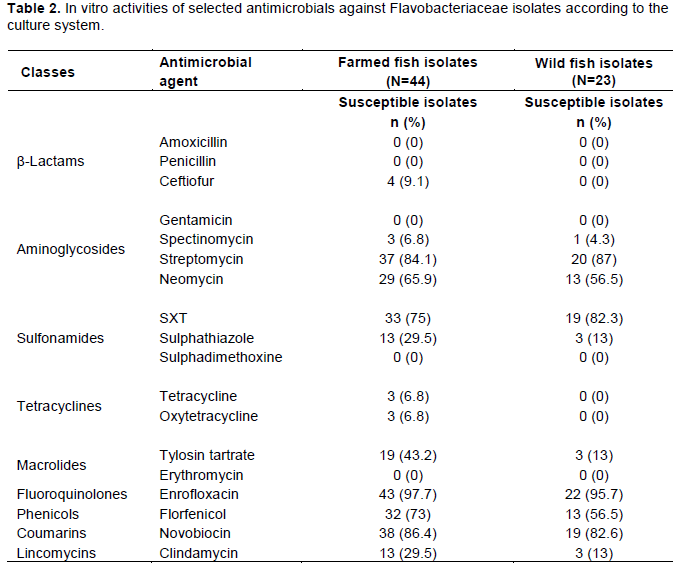
The in vitro activity of the antimicrobial agents was also evaluated based on farmed and wild fish and sampling regions. Generally, susceptibility levels among isolates did not vary significantly between neither farmed versuswild fish nor sampling region. One observation was, however, that while none of the isolates from wild fish were susceptible to ceftiofur and tetracycline/ oxytetracycline, respectively three and four isolates from farmed fish were susceptible to these antimicrobials (Table 2).
Multiple antimicrobial resistance (MAR) index
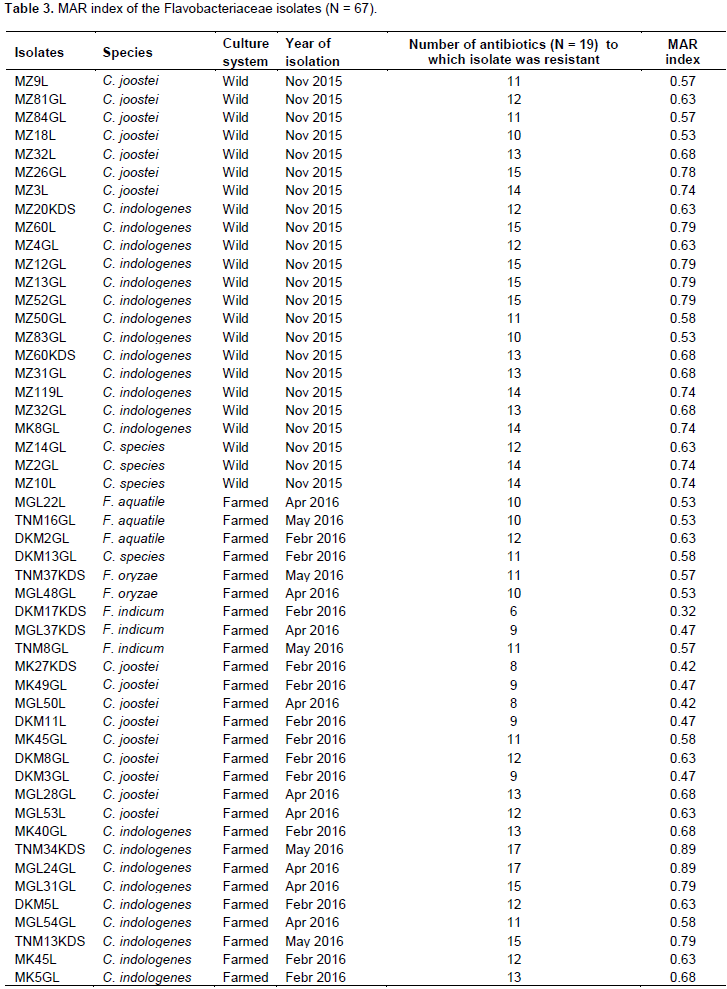
Overall antimicrobial susceptibility pattern of each isolate is also summarized in Table 3. The susceptibility patterns varied within groups of isolates from the same site or sample, and also within the same species. Most of the C. indologenes isolates showed low susceptibility up to 17 of the 19 antimicrobials tested, which was the highest degree of multi-resistance observed in this study regardless of sample origin. The lowest level of resistance was observed in F. indicum, which was sensitive to 6 of 19 antimicrobials tested. Moreover, C. indologenes and C. joostei, which were isolated from almost all the sampling sites/fish, showed a broader range of MAR indexes. The highest MAR index was 0.89 observed in C. indologenes and the lowest was 0.32 observed in C. indicum isolates, indicating that none of the sampling sites could be characterized as low-risk sources of contamination. Comparison of the different culture system showed that all isolates recovered from wild and farmed Nile tilapia revealed multiple resistance to all antimicrobials tested.
Antimicrobial resistance has become a global health threat involving the environmental, animal and human sectors. Knowledge of antimicrobial susceptibility and resistance patterns in microbes from many biological compartments is important to understand the development and transmission of resistance within and between bacterial reservoirs as well as identifying treatment alternatives against specific infections. In this study, we report data on antimicrobial susceptibility of Flavobacteriaceae isolated from wild and farmed Nile tilapia in Tanzania. Our results show variable antimicrobial susceptibility patterns against 19 antimicrobial agents commonly used in veterinary medicine and aquaculture in most developed countries.
This is in concordance with previous studies showing that Flavobacteriaceae isolates are able to grow in high concentrations of several antimicrobial agents, suggesting the presence of intrinsic or acquired resistance mechanisms, resulting in reduced susceptibility to multiple antimicrobial agents (Akinbowale et al., 2006, Clark et al., 2009, Henríquez-Núñez et al., 2012, Hesami et al., 2010).
All isolates were resistant to the β-lactams amoxicillin and penicillin, to the aminoglycoside gentamycin, the macrolide erythromycin and to sulfadimethoxine. Resistance to β-lactams was expected as the commonly chromosomally encoded Amber Class B and other β-lactamase enzymes have been previously broadly recognized in Flavobacteriaceae (Bellais et al., 2002b, Bellais et al., 2002a; Gonzalez and Vila, 2012). Resistance to erythromycin is also consistent with previous studies, in which MICs of > 32 μg/ml have been reported amongst Flavobacteriaceae (Clark et al., 2009, Darwish et al, 2008, Declercq et al., 2013). In contrast to the present investigation, a previous study reported 100 % susceptibility (MIC90 >8μg/ml) to gentamycin in Flavobacterium columnare isolates collected worldwide from 17 fish species (Declercq et al., 2013).
Most (97%) Flavobacteriaceae isolates were susceptible to fluoroquinolone enrofloxacin. The variable levels of susceptibility found for streptomycin, novobiocin and sulfamethoxazole/ trimethoprim tested in this study, suggest that they may be useful for treatment of infections caused by some strains. However, sensitivity testing should be performed against the actual disease strain before any of these antimicrobials are considered for use. Variation in antimicrobial susceptibility amongst Flavobacteriaceae and other fish bacteria have been reported elsewhere (Akinbowale et al., 2006, Schmidt et al., 2000). In general, Flavobacteriaceae are recognized as intrinsically resistant to several antimicrobial agents, and only a few specific transferable resistance determinants known from other bacterial genera have been identified. Several genes coding for efflux pumps have been associated with intrinsic resistance in Flavobacteriaceae isolates. For example, previous study investigated genome sequence of Flavobacterium johnsoniae and revealed presence of chloramphenicol inducible multidrug efflux pump system of RND family (Clark et al., 2009, Henríquez-Núñez, 2012, Michel et al., 2005). In addition, the low susceptibility of Flavobacteriaceae to florfenicol has been associated with expression of multidrug efflux pumps (Michel et al., 2005). Moreover, it has been demonstrated that efflux pumps are important for other processes such as detoxification of intracellular metabolites, bacterial virulence, cell homeostasis and intercellular signal trafficking (Martinez et al., 2009; Pasqua et al., 2019).
Most isolates in the present study were inhibited by low concentrations of florfenicol, but since 37% were able to grow in higher florfenicol concentrations, the presence of acquired resistance mechanisms cannot be ruled out. The banning of use of phenicol, due to its adverse effect in humans, has probably aided in reducing the risk of acquired resistance in aquaculture environments (Akinbowale et al., 2006; Tsai et al., 2019). However, other studies have insisted that monitoring resistance of florfenicol is of great importance as it is the main antimicrobial agent used to treat infections caused by Flavobacteriaceae (Verner-Jeffreys et al., 2015). Unfortunately, no information regarding the use of florfenicol or other phenicol compounds was available in the sampling areas.
Tetracycline and/or oxytetracycline have been frequently used in fish farming, particularly to control systemic bacterial infections of fish in most developed countries (Miranda et al., 2018; Jerbi et al., 2011), and it has been found that the continued and widespread use of tetracycline has led to the development of resistant bacteria in all aspects of fish farming (Higuera-Llantén et al., 2018; Mirand and Zemelman, 2002). This study revealed low susceptibility to tetracycline and/or oxytetracycline, with MIC ranges of 0.25 - 8 μg/ml. Other workers recorded considerable frequencies of tetracycline resistance amongst Flavobacteriaceae isolated from aquaculture environments and associated it with imprudent use of antimicrobials and acquired resistance (Schmidt et al., 2000). However, due to lack of information relating to the treatment history of the sampled fish, it is difficult to associate the low susceptibility observed with acquired resistance or imprudent use of antimicrobials.
All Flavobacteriaceae were found with MAR index values greater than 0.2 in this study. Previous studies have shown that bacteria originating from an environment where several antimicrobials are used usually display MAR indexes greater than 0.2 (Tambekar et al., 2006). Thus, the fact that all MAR indexes recorded were above 0.2 indicate that they are from the high risk source where antibiotics are frequently used, possibly from surrounding study areas. Furthermore, low susceptibility results in this study concur with the findings of previous studies (Clark et al., 2009), who found multidrug resistance in Flavobacteria and Chryseobacterium species isolated from fish. Nile tilapia is the most cultured and widely consumed fish species in Tanzania. Lake Victoria is the major capture fish source in the country and also is a source for tilapia fingerlings to most fish farmers in Tanzania. The assessment of Antimicrobial resistance (AMR) is of great importance as dissemination of AMR bacteria to fish could lead to serious public health risks. Interviews conducted in our earlier study did not reveal a history of antimicrobial use in farmed fish during the present study (Mwega et al., 2018). Our results suggest that isolates from wild fish were slightly more resistant against the antimicrobials than the isolates from farmed fish. Although the difference is not significant among groups, this observation could be explained by the fact that the wild Nile tilapia was captured at the shorelines of Lake Victoria which makes the two environments more alike. Systematic data on use of antimicrobials to the ponds are not available in Tanzania, so we do not know how much and how frequent antimicrobials are used. Therefore, it is not possible to draw any conclusions on the association between antimicrobials use and the levels of resistance observed in the present study.
In this study, we have found that Flavobacteriaceae isolates are resistant to several antimicrobials tested. The ability of Flavobacteriaceae to resist multiple drugs is indeed was expected, as it is known that most of these isolates are intrinsically resistant to several classes of antimicrobials and/ or can acquire resistant genes from other environmental microbes. However, whether the antimicrobial resistance traits are acquired through gene transfer or intrinsic is not clearly elucidated at the moment. This can be determined by analyzing whole genomes of those isolates in the future. The findings from this study give us insight into appropriate choice of antimicrobial agents for effective treatment of infections caused by these isolates.
The authors have not declared any conflict of interests.
The authors appreciate Mr. Jeremiah Mgusi, Ms. Mirium Makange (SUA), Kristin O’Sullivan and Helge Christoffer Høgberg Hansen (NMBU) for their technical support and also thankful to Genome Science Centre at CVMB - SUA for laboratory facilities. The authors are grateful for the financial support provided to the TRAHESA project by the Norwegian Agency for Development Cooperation (NORAD) through the Norwegian Programme for Capacity Building in Higher Education and Research for Development (NORHED), project number Tan-13/0027.
REFERENCES
|
Akinbowale OL, Peng H, Barton MD (2006). Antimicrobial resistance in bacteria isolated from aquaculture sources in Australia. Journal of Applied Microbiology 100(5):1103-1113.
Crossref
|
|
|
|
Bellais S, Naas T, Nordmann P (2002a). Molecular and biochemical characterization of Ambler class A extended-spectrum β-lactamase CGA-1 from Chryseobacterium gleum. Antimicrobial Agents Chemotherapy 46(4):966-970.
Crossref
|
|
|
|
|
Bellais S, Naas T, Nordmann P (2002b). Genetic and biochemical characterization of CGB-1, an Ambler class B carbapenemhydrolyzing β-lactamase from Chryseobacterium gleum. Antimicrobial Agents Chemotherapy 46(9):2791-2796.
Crossref
|
|
|
|
|
Bernardet JF, Bowman JP (2011). Genus I. Flavobacterium Bergey et al. 1923. In Bergey's Manual of Systematic Bacteriology, 2nd edn, vol. 4, pp. 112-154. Edited by W. Whitman. Baltimore: The Williams & Wilkins Co., Baltimore
|
|
|
|
|
Bernardet JF, Nakagawa Y (2006). An introduction to the family Flavobacteriaceae. In: Dworkin M, Falkow S, Rosenberg E, Schleifer KH, Stackebrandt E, editors. The prokaryotes: Delta, Epsilon Subclass. Springer New York7:455-480.
Crossref
|
|
|
|
|
Cabello FC (2006). Heavy use of prophylactic antibiotics in aquaculture, a growing problem for human and animal health and for the environment. Environmental Microbiology 8(7):1137-1144.
Crossref
|
|
|
|
|
Cabello FC, Godfrey HP, Tomova A, Ivanova L, Dölz H, Millanao A (2013). Antimicrobial use in aquaculture re-examined, its relevance to antimicrobial resistance and to animal and human health. Environmental Microbiology 15(7):1917-1942.
Crossref
|
|
|
|
|
Clark SE, Jude BA, Danner GR, Fekete FA (2009). Identification of a multidrug efflux pump in Flavobacterium johnsoniae. Veterinary Research 40(6):1-10.
Crossref
|
|
|
|
|
Clinical and Laboratory Standards Institute (CLSI) (2014). Methods for Broth Dilution Susceptibility Testing of Bacteria Isolated from Aquatic Animals, Approved Guideline VET04-A2. Clinical and Laboratory Standards Institute. Wayne, PA.
|
|
|
|
|
Darwish AM, Farmer BD, Hawke JP (2008). Improved method for determining antibiotic susceptibility of Flavobacterium columnare isolates by broth microdilution. Journal of Aquatic Animal Health: 20(4):185-191.
Crossref
|
|
|
|
|
Darwish AM, Ismaiel AA, Newton JC, Tang J (2004). Identification of Flavobacterium columnare by a species-specific polymerase chain reaction and renaming of ATCC43622 strain to Flavobacterium johnsoniae. Molecular and cellular probes 18(6):421-427.
Crossref
|
|
|
|
|
Declercq AM, Boyen F, Van Den Broeck W, Bossier P, Karsi A, Haesebrouck F, Decostere A (2013). Antimicrobial susceptibility pattern of Flavobacterium columnare isolates collected worldwide from 17 ï¬sh species. Journal of Fish Diseases 36(1):45-55 .
Crossref
|
|
|
|
|
Gonzalez LJ, Vila AJ (2012). Carbapenem resistance in mediated by metallo-β-lactamase BlaB. Antimicrobial Agents Chemotherapy 56(4):1686-1692.
Crossref
|
|
|
|
|
Gufe C, Canaan Hodobo T, Mbonjani B, Majonga O, Marumure J, Musari S, Machakwa J (2019). Antimicrobial Profiling of Bacteria Isolated from Fish Sold at Informal Market in Mufakose, Zimbabwe. International Journal of Microbiology.
Crossref
|
|
|
|
|
Henríquez-Núñez H, Evrard O, Kronvall G (2012). Antimicrobial susceptibility and plasmid profiles of Flavobacterium psychrophilum strains isolated in Chile. Aquaculture 354:38-44.
Crossref
|
|
|
|
|
Hesami S, Parkman J, Macinnes JI (2010). Antimicrobial susceptibility of Flavobacterium psychrophilum isolates from Ontario. Journal of Aquatic Animal Health 22(1):39-49.
Crossref
|
|
|
|
|
Higuera-Llantén S, Vásquez-Ponce F, Barrientos-Espinoza B, Mardones FO, Marshall SH, Olivares-Pacheco J (2018). Extended antibiotic treatment in salmon farms select multiresistant gut bacteria with a high prevalence of antibiotic resistance genes. PLoS ONE 13(19).
Crossref
|
|
|
|
|
Izumi S, Aranishi F (2004). Relationship between gyrA mutations and quinolone resistance in Flavobacterium psychrophilum isolates. Applied Environmental Microbiology 70(7):3968-3972.
Crossref
|
|
|
|
|
Jensen LB, Angulo FJ, Molbak K, Wegener HC (2008). Human health risks associated with antimicrobial use in animals. Guide to Antimicrobial Use in Animals pp. 13-26.
Crossref
|
|
|
|
|
Jerbi MA, Ouanes Z, Besbes R, Achour L, Kacem A (2011). Single and combined genotoxic and cytotoxic effects of two xenobiotics widely used in intensive aquaculture. Mutation Research 724(1-2):22-27.
Crossref
|
|
|
|
|
Kumru S, Tekedar HC, Gulsoy N, Waldbieser GC, Lawrence ML, Karsi A (2017). Comparative Analysis of the Flavobacterium columnare Genomovar I and II Genomes. Frontiers in Microbiology 8:1375.
Crossref
|
|
|
|
|
Loch TP, Faisal M (2015). Emerging flavobacterial infections in fish: A review. Journal of Advanced Research 6(3):283-300.
Crossref
|
|
|
|
|
Loch TP, Fujimoto M, Woodiga SA, Walker ED, Marsh TL, Faisal M (2013). Diversity of fish-associated Flavobacteria of Michigan. Journal of Aquatic Animal Health 25(3):149-64.
Crossref
|
|
|
|
|
Madsen L, Dalsgaard I (2000). Comparative studies of Danish Flavobacterium psychrophilum isolates: ribotypes, plasmid profiles, serotypes, and virulence. Journal of Fish Diseases 23(3):211-218.
Crossref
|
|
|
|
|
Martinez JL, S'anchez MB, Mart'ınez-Solano L (2009). Functional role of bacterial multidrug efflux pumps in microbial natural ecosystems. FEMS Microbiology Review 33(2):430-449.
Crossref
|
|
|
|
|
McBride MJ (2014). The Family Flavobacteriaceae. In: Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F. (eds) The Prokaryotes. Springer, Berlin, Heidelberg.
|
|
|
|
|
Michel C, Matteâ€Tailliez O, Kerouault B, Bernardet JF (2005). Resistance pattern and assessment of phenicol agents' minimum inhibitory concentration in multiple drug resistant Chryseobacterium isolates from fish and aquatic habitats. Journal of applied microbiology 99(2):323-332.
Crossref
|
|
|
|
|
Mirand CD, Zemelman R (2002). Antimicrobial multiresistance in bacteria isolated from freshwater Chilean salmon farms. The Science of the total environment: 293(1-3):207-218.
Crossref
|
|
|
|
|
Miranda CD, Godoy FA, Lee MR (2018). Current Status of the Use of Antibiotics and the Antimicrobial Resistance in the Chilean Salmon Farms. Frontiers in microbiology 9:1284.
Crossref
|
|
|
|
|
Mwega E, Colquhoun D, Tuntufye H, Mdegela R, Mutoloki S, Evensen Ø, Wasteson Y (2018). Isolation and Characterization of Flavobacteriaceae from Farmed and Wild Nile Tilapia in Tanzania. Journal of Aquatic Animal Health 31(1):23-30.
Crossref
|
|
|
|
|
Pasqua M, Grossi M, Zennaro A, Fanelli G, Micheli G, Barras F, Colonna B, Prosseda G (2019). The Varied Role of Efflux Pumps of the MFS Family in the Interplay of Bacteria with Animal and Plant Cells. Microorganisms 7(9):285.
Crossref
|
|
|
|
|
Pathmalal M (2018). Heavy use of antibiotics in aquaculture: Emerging human and animal health problems - A review. Sri Lanka Journal of Aquatic Science 23:13-27.
Crossref
|
|
|
|
|
Pilarski F, Rossini AJ, Ceccarelli PS (2008). Isolation and characterization of Flavobacterium columnare (Bernardet et al. 2002) from four tropical fish species in Brazil. Brazillian Journal of Biology 68(2):409-414.
Crossref
|
|
|
|
|
Ratnamani M, Rao R (2013). Elizabethkingia meningoseptica: Emerging nosocomial pathogen in bedside hemodialysis patients. Indian Journal of Critical Care Medicine 17(5):304-307.
Crossref
|
|
|
|
|
Samuel L, Marian MM, Apun K, Lesley MB, Son R (2011). Characterization of Escherichia coli isolated from cultured catfish by antibiotic resistance and RAPD analysis. International Food Research 18(3):971-976.
|
|
|
|
|
Schmidt AS, Bruun MS, Dalsgaard I, Pedersone K, Larsen, JL (2000). Occurrence of antimicrobial resistance in fish-pathogenic and environmental bacteria associated with four Danish rainbow trout farms. Applied Environmental Microbiology 66(11):4908-4915.
Crossref
|
|
|
|
|
Tambekar DH, Dhanorkar DV, Gulhane SR, Khandelwal VK, Dudhane MN (2006). Antibacterial susceptibility of some urinary tract pathogens to commonly used antibiotics. African Journal of Biotechnology 5(17):1562-1565.
|
|
|
|
|
Tsai MY, Lin CF, Yang WC, Lin CT, Hung KH, Chang GR (2019). Health risk assessment of banned veterinary drugs and quinolone residues in shrimp through liquid chromatography-tandem mass spectrometry. Applied Sciences 9(12):2463.
Crossref
|
|
|
|
|
Verner-Jeffreys D, Taylor NGH, Pitlochry, Perthshire (2015). Review of Freshwater Treatments Used in the Scottish Freshwater Rainbow Trout Aquaculture Industry. Scottish Aquaculture Research Forum Report SARF100 Available @:
View
|
|
|
|
|
Watts JEM, Schreier HJ, Lanska L, Hale MS (2017). The Rising Tide of Antimicrobial Resistance in Aquaculture: Sources, Sinks and Solutions. Marine Drugs 15(6):158.
Crossref
|
|