ABSTRACT
This study determines the antibiotic resistance pattern of extended spectrum beta-lactamase (ESBL) producing pathogens responsible for catheter associated urinary tract infection (CAUTI) and the genes associated with the resistance. The study used 35 ESBL-producing pathogens isolated from urine and biofilm found in CAUTI from June 2018 to November 2018. Pathogens were confirmed phenotypically for ESBL production with cefpodoxime combination disc kits. Antibiotic resistance was tested using the Kirby-Baurer disc diffusion method on Muller Hinton Agar. ESBL genotypes were identified with PCR. Urine isolates showed higher frequency of resistance against ciprofloxacin (94.11%), cefuroxime and ceftazidime (82.35%) and with no recorded resistance against Eertepenem (0.0%). The average resistance of the biofilm isolates ranged from 0.0% (Ertepenem) to 88.89% (Cefuroxime, Cefpodoxime, Ciprofloxacin and Trimethoprim). All the targeted genes were identified with CTX-M (40%) being the most dominant among them. The ESBL-producing pathogens showed zero resistance against Ertepenem. Ciprofloxacin and other Cephems commonly used in CAUTI treatment were shown to be less effective. The high resistance is as a result of the bacterial cells present in the biofilm with Klebsiella pneumonia exhibiting more resistance than the ESBL-producing E. coli and CTX-M-1 was identified as the most prevalent gene among the identified genotypes. Ertepenem should therefore be recommended for treatment of Catheter associated urinary tract infection.
Key words: Catheter, CAUTI, extended spectrum beta-lactamase (ESBL), urine, biofilm, resistance, microorganisms.
The incidence of Catheter Associated Urinary Tract Infection (CAUTI) is inevitable having in mind that there is about 10% daily infection rate when catheter is present
in situ (Hartley and Valley, 2014; Hooton et al., 2010). This infection can lead to more serious complications such as sepsis and endocarditis, and it is estimated that over 13000 deaths each year are associated with healthcare-associated UTIs in the United States of America (Parida and Mishra, 2013). The microorganisms responsible for this infection are mostly the micro flora from the patient, healthcare attendants and the environment. These organisms are able to form biofilm when they attach themselves on these solid surfaces for extended periods of time. Microorganisms usually encountered in CAUTI include
Escherichia coli, Klebsiella pneumonia, other enterobacteriaceae and
Staphylococcus species (Albu et al., 2018; Percival et al., 2018).
E. coli and
K. pneumonia are the leading cause of CAUTI with increasing resistant rates (
Köves et al., 2017; Percival et al., 2018).
These microorganisms have the ability to hydrolyze both beta lactam antibiotics (3rd generation cephalosporins, penicillins and cephamycins) and non-beta lactam antibiotics such as tetracycline, chloramphenicol, aminoglycosides, quinolones as well as vancomycin (Shaikh et al., 2015). This may be attributed to the overuse of antibiotics for infection treatments, the ability of the microbes to grow as biofilms, genetic mutations and pathogens sharing resistance genes with each other (Albu et al., 2018). The extended spectrum beta-lactamase (ESBL) enzymes are classified into four (A, B, C and D) variants based on their amino acid sequences. The most common class, A β-lactamases, encountered in K. pneumonia and E. coli are the SHV (Sulphydryl variable), TEM (Temoniera) and CTX-M (cefotaximase). Both SHV and TEM are plasmid mediated, degrading penicillins and first generation cephalosporins. These enzymes are however inhibited by clavulanic acid but susceptible to third generation cephalosporins (Paterson et al., 2001). CTX-M (cefotaximase), is another class A ESBL enzyme which preferentially hydrolyzes cefotaxime as compared to ceftazidime and it is organized in major groups as CTX-M- (1, 2, 8, 9 and 25). Reid et al. (2018) reports that CTX-M is increasing and spreading faster and therefore becoming more prevalent than TEM and SHV. Oxacillinase (OXA), a class D ESBL with the ability to hydrolyze oxacillin, is mostly responsible for carbapenems resistance but it is not as common as TEM, SHV and CTX-M (Dallenne et al., 2010).
Several studies described specific genotypes in the ESBL producing pathogens causing resistance to some antibiotics globally (Reid et al., 2018; Sana et al., 2011; Bajpai et al., 2017; Lewis et al., 2007). In India TEM has been reported to be frequent among urine isolates (Bajpai et al., 2017) whilst in Europe, United States and Africa, CTX-M is mostly detected (Reid et al., 2018; Lewis et al., 2007; Zeynudin et al., 2018). In Ghana the prevalence of ESBL reported from the major teaching hospitals ranges from 37.96% to 57.8% with TEM and CTX-M variants being prevalent among E. coli and K. pneumonia (Mensah et al., 2016; Agyekum et al., 2016). This is of major concern, since both pathogens are the predominant cause of health and community related infections. Even though some studies have investigated ESBL genes causing antibiotic resistance in Ghana (Agyekum et al., 2016; Feglo and Adu, 2016; Hackman et al., 2014), there is no data on antibiotic resistance of ESBL producing pathogens related to CAUTI in Ghana. This study investigates the resistance pattern of ESBL-producing pathogens causing CAUTI and the genes associated with such resistance using the polymerase chain reaction method as a novel way to bridge the gap by providing baseline surveillance on CAUTI, it prevalence and antimicrobial resistance pattern at Komfo Anokye Teaching Hospital (KATH) in Ghana.
Study site
Urine and biofilm samples were collected from 105 catheterized patients at the urology unit of Komfo Anokye Teaching Hospital (KATH), Kumasi from June 2018 to November 2018. The hospital is a 1200 bed capacity facility located in the heart of Kumasi, the capital town of the Ashanti region. It is the second largest referral/teaching hospital in the country. It serves 12 out of the 16 administrative regions of the country and neighboring countries like Burkina Faso, Togo and Ivory Coast. This is due to the hospital’s location and the road network of the country.
Sampling
All samples tested positive for CAUTI. Criteria for CAUTI were based on catheterization duration, bacteria load and patients’ symptoms as described by CDC/IDSA. All samples that met CAUTI definition due to ESBL producing E. coli and K. pneumonia were used for further tests.
The ESBL producing isolates stored in brain heat infusion and glycerol broth were sub cultured on nutrient agar (Oxoid, UK) and incubated at 37°Ï¹ for 24 h. The biofilms from the catheters were pre enriched with 15 ml of maximum recovery diluent and shaken for 2 min to extract biofilm from the catheter lumen into the diluent and incubated at 37°C for about 6 h before inoculation. Morphological appearance, Gram staining and other standard biochemical methods were used to confirm the identity of the ESBL isolates. ESBL detection was phenotypically confirmed with the Cefpodoxime combination disc kits (Oxiod, UK) using the disc diffusion method on Muller-Hinton Agar (MHA) (Oxiod, UK) as described by CLSI (2020). Zone of diameter ≥5 mm observed in the combined disc when compared to the single disc was considered positive ESBL production. Quality control was done using K. pneumonia ATCC 700603 and E. coli ATCC 25922 organisms.
Antibiotic resistance test
The antibiotic resistance test was done on the ESBL-producing isolates using the disc diffusion method on MHA with the following antibiotics; Ertapenem (10 μg), Cefuroxime (30 μg), Cefpodoxime (30 μg), Ceftazidime (30 μg), Fosfomycin (50 μg), Ciprofloxacin (5 μg), Cefazolin (30 μg), Cefoxitin (30 μg), Trimethoprim (5 μg) and Nitrofurantoin (300 μg). Zones of inhibition around the discs were measured and compared with the CLSI break points for interpretation.
DNA extraction
Using the boiling method of DNA extraction, two to three bacteria colonies from nutrient agar were inoculated in 200 μl nuclease free water. The suspension was placed in a 100°Ï¹ heat block for 10 minand centrifuged for 10 min at a speed of 14000 rpm. The supernatant was pipetted into a new tube for the PCR analysis.
Gene identification using multiplex PCR
Multiplex PCR was carried out with bla TEM, bla SHV and bla OXA primers in a single tube. The master mix prepared was in 50 μl reaction volume that contained 25 μl of Emerald premix (2×), 7.5 μl primer mix, 15.5 μl of nuclease free water and 2 μl of the DNA template. DNA amplification was done in a thermo cycler. Table 1 shows the cycling conditions and primer sequence.
A separate multiplex PCR was carried out for bla CTX-M-1, CTX-M-2 and CTX-M-9 genes. The multiplex reaction was performed in 50 μl reactions containing; 25 μl of 2×multiplex PCR buffer with Mg²+ and dNTP plus, 0.25 μl multiplex PCR enzyme mix, 13.75 μl of nuclease free water, 2 μl of DNA template and primers listed in Table 2 with their reaction volumes, sequences, target gene, cyclic condition and product size. Multiplex PCR was run on 2% agarose gel and 1×TAE buffer at 100 V for 30 min. Gel bands were viewed using a gel transluminator (Reid et al., 2018).
Data analysis
Data was entered into a Microsoft excel and analyzed using graph Pad Prism 5. Study variables were subjected to descriptive analysis such as frequencies and percentages. Chi-square goodness of fit examine was run with chisq.test function in the stats package in R programme (version 4.0.0; R Core Team, 2020) to assess the difference in resistance of E. coli and K. pneumonia under each antibiotic. A value of p Ë‚ 0.05 was regarded as statistically significant.
Ethical approval
Ethical clearance was obtained from the committee on Human Research, Publication and Ethics of the Kwame Nkrumah University of Science and Technology, Kumasi, Ghana. Written informed consent was obtained from all study participants. Infected patients were treated based on their laboratory result.
Antibiotic resistance of ESBL- producing pathogens isolated from urine specimen
The ESBL producing pathogens isolated from the urine show high resistance to Ciprofloxacin (94.11%) followed by the four Cephems/beta-lactam antibiotics with 82.35% for Cefuroxime and Ceftazidime and 76.47% for Cefpodoxime and Cefazolin. Comparing the resistance between K. pneumonia and E. coli, two of the antibiotics (Fosfomycin and Trimethoprim) were found to be more resistant against E. coli than K. pneumonia. However, Cefpodoxime (p>0.05) was not significantly different from K. pneumonia (Table 3). For Ertepenem no resistance was found for the ESBL-pathogens. On average lower resistance was recorded for Fosfomycin (17.64%), Cefoxitin (35.49%) and Nitrofurantoin (41.18%).
Antibiotic resistance of ESBL- producing pathogens isolated from biofilm specimen
For the biofilm isolates, the average resistance numbers ranged between 38.88% (Cefoxitin) and 88.89% (Cefuroxime, Cefpodoxime, Ciprofloxacin and Trimethoprim). There was statistical difference between E. coli and K. pneumonia for all tested drugs except for Ertepenem. E. coli was significantly more resistant than K. pneumonia to Ciprofloxacin (0.000) and Cefoxitin (0.008). Apart from Cefoxitin and Ertepenem, all the K. pneumonia were resistant to the rest of the antibiotics. The resistance pattern as presented in Table 4 shows that K. pneumonia was more resistant than E. coli. However, none of the ESBL-pathogen from the biofilm recorded resistance against Ertepenem.
PCR detection of ESBL genes
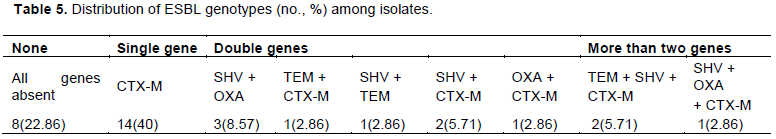
Thirty-five ESBLs were analyzed using the multiplex PCR to identify specific resistance genes such as TEM, SHV, OXA and CTX-M. The CTX-M genes were further analyzed to identify CTX-M-1, CTX-M-2 and CTX-M-9 subgroups. More than half of the resistance genes identified were CTX-M, followed by SHV, OXA and the TEM. The 23 isolates identified to contain CTX-M genes were sub grouped into three by multiplex PCR analysis. These isolates had at least one of the sub groups analyzed. CTX-M-1 was most prevalent 14 (60.87%) followed by 8 (34.78%) for CTX-M-9 and 1 (4.35%) for CTX-M-2. The total prevalence of resistance genes identified in the isolates is 77.14%. CTX-M was only identified in 40% of the isolates, 5 double genes were identified in 28.57% isolates (SHV+ OXA, SHV + TEM, SHV + CTX –M, TEM + CTX-M, OXA+ CTX –M), and three combined genes were identified in 8.57% isolates (TEM + SHV + CTX-M, SHV + OXA + CTX-M). None of the resistant genes was detected in 22.86% of the isolates. Table 5 shows that, at least one of the resistance genes was identified in each of the isolates. However, isolates that had two or three resistance genes mostly recorded CTX-M and SHV as part of the combined resistance genes.
ESBL-producing pathogens isolated from urine
Resistance of ESBL-producing K. pneumonia and E. coli to ciprofloxacin in this study is similar to what was reported by Hyun et al. (2015) who recorded about 80% resistance among ESBL-producing K. pneumonia and E. coli. The resistance rate of E. coli for this study was also similar to those of earlier reported study (Albu et al., 2018). It was also established that, four out of the five tested Cephems/beta-lactam antibiotics were resistant confirming a previous study in Ghana (Agyekum et al., 2016) and in France (Martin et al., 2016). High resistance against fluroquinolones and β-lactam in this present study could be due to excessive use of such antibiotics in the UTI therapy. This is of major concern since this group of antibiotics are frequently prescribed and consumed in our setting (Afriyie et al., 2018). On the other hand, none of the isolates recorded a resistance to Ertepenem which is in agreement with other studies in Ghana indicating no Carbapenem resistance (Agyekum et al., 2016; Hackman et al., 2014). This affirms the fact that Ertepenem and other Carbapenems are not prescribed routinely in Ghana according to the guidelines of the country’s standard health treatment even though Carbapenem resistance has recently been recorded in other parts of Africa (Okoche et al., 2015).
Other drugs with low resistance recorded in this study were Fosfomycin, Cefoxitin and Nitrofurantoin which is in conformity to other similar studies (Sana et al., 2011; Hyun et al., 2015). This finding could be associated with less accessibility and high price of these drugs in the local market as suggested by Albu et al. (2018) and Feglo and Adu (2016) when they identified K. pneumonia as more resistance than E. coli, while a study in Korea reported otherwise (Hyun et al., 2015).
ESBL-producing pathogens isolated from biofilm
The ESBL
-producing
K. pneumonia isolated from biofilm showed resistance to all Nitrofurans, Fosfomycins, Fluoroquinolones, Folate pathway and all but one Cephems which is in accordance with a study by Vuotto et al. (2017) in which ESBL uropathogens were resistant to similar classes of antibiotic. Our results are also supported by a study that suggested that genes responsible for antibiotic resistance correlates with genes involved in biofilm production (Subramanian et al., 2012). In other studies there was 95% tendency of ESBL-producing
E. coli to form biofilm than for non ESBL-producing
E. coli. This might explain why
E. coli showed high resistance to all antibiotics except the Carbapenem (Ertepenem) (Neupane et al., 2016; Subramanian et al., 2012). Biofilm protects bacterial cells against the effect of antibiotica resulting in treatment failure also for new and powerful drugs (
Verderosa et al., 2019). Another treatment difficulty may occur with the ability of the bacteria to produce several ESBL enzymes simultaneously.
That the ESBL-producing pathogens resistance is not limited to Cephem/beta-lactam class of antibiotics indicates the ability for cross-resistance to other classes of antibiotics as has been previously observed in some studies in Ghana, limiting the therapeutic option for complicated infections (Feglo and Adu 2016; Hackman et al., 2014). The resistance of ESBL-producing pathogens isolated from biofilm to Ciprofloxacin is consistent with earlier studies (Subramanian, 2012; Neupane et al., 2016). However, Ertepenem, Cefoxitin and Nitrofurantoin were effective against the biofilm producing pathogens, which is in agreement with earlier findings in Nepal and India (Neupane et al., 2016; Shanmugam et al., 2017). The biofilm isolates were more resistant than the urine isolates. The K. pneumonia higher resistance pattern compared to E. coli is in agreement with some earlier studies (Albu et al., 2018; Feglo and Adu, 2016) but contrary to the findings of Hyun et al. (2015). Even though Ertepenem was active for both biofilm and urine isolates the results contradicts that of Vuotto et al. (2017).
Molecular detection of ESBL genes
Resistance genes in the Enterobacteriaceae were specifically identified using the multiplex PCR, with r the high prevalence rate of 53.48% to CTX-M. This result is in accordance with a studies in Leicestershire, UK (Reid et al., 2018), Ghana (Agyekum et al., 2016) and Nigeria (Nuhu et al., 2020) but in contrast to a similar study in India (Bajpai et al., 2017) were TEM was the predominate gene i. These notwithstanding, other studies in Ghana and elsewhere outline CTX-M-15 as the dominant genotype in the CTX-M group (Lewis et al., 2007; Agyekum et al., 2016; Eibach et al., 2018; Reid et al., 2018; Hackman et al., 2014). It is worth noting that CTX-M-1 are prevalent in domestic animals like cats and dogs (Ghazanfar et al., 2019) as well as in both local and imported poultry products (Eibach et al., 2018). The CTX-M-1 prevalence found in this study could be attributed to the common lifestyle of many Ghanaian sharing their compound with dogs and cats as food or pet coupled with high consumption of poultry products reported to contain the CTX-M-1 genotype (Kwadzo et al., 2015). About 60% of the isolates had multi-ESBL genotypes with some of their combinations being (CTX +TEM, SHV + TEM and CTX+TEM +SHV) which is consistent with a previous study at the same facility (Feglo and Adu 2016). Of all the seven multi-ESBL genotypes identified, CTX-M and SHV were mostly present in each combination. This gives credence to CTX-M and SHV as key components to antibiotic resistance.
It is clear that CAUTI caused by ESBL-producing E. coli and K. pneumonia are highly resistant against commonly used antibiotics. Since isolates from the urine also show resistance to many of the antibiotics that were surveyed, this high resistance may partially be credited to the bacteria cells in the biofilm with K. pneumonia exhibiting more resistance than the ESBL-producing E. coli. Ciprofloxacin and other Cephems commonly used in CAUTI treatment were less effective. However, Ertepenem which recorded no bacteria resistance to the biofilm isolates, could be considered as the best drug of choice for CAUTI treatment. The one resistant genotype identified in about 80% of the ESBL-pathogens was CTX-M-1 and it is the most prevalent gene among the identified genotypes. Ertepenem and other Carbapenems should be considered for treatment of CAUTI since they also eliminate biofilm pathogens.
The authors have not declared any conflict of interests.
REFERENCES
|
Afriyie DK, Adu LB, Dzradosi M, Amponsah SK, Ohene-Manu P, Manu-Ofei F (2018). Comparative in vitro activity of ciprofloxacin and levofloxacin against isolated uropathogens in Ghana: a pilot study. Pan African Medical Journal 30:194.
Crossref
|
|
|
|
Agyekum A, Fajardo-Lubián A, Ansong D, Partridge SR, Agbenyega T, Iredell JR (2016). blaCTX-M-15 carried by IncF-type plasmids is the dominant ESBL gene in Escherichia coli and Klebsiella pneumoniae at a hospital in Ghana. Diagnostic Microbiology Infectious Diseases 84(4):328-33.
Crossref
|
|
|
|
|
Albu S, Voidazan S, Bilca D, Badiu M, Trut A, Ciorea M (2018). Bacteriuria and asymptomatic infection in chronic patients with indwelling urinary catheter. Medicine (Baltimore) 97(33):e11796.
Crossref
|
|
|
|
|
Bajpai T, Pandey M, Varma M, Bhatambare GS (2017). Prevalence of TEM, SHV, and CTX M Beta-Lactamase genes in the urinary isolates of a tertiary care hospital. Avicenna Journal of Medicine 7(1):12-16.
Crossref
|
|
|
|
|
CLSI (2020). M100 Performance Standards for Antimicrobial Susceptibility Testing. 30th Ed.
|
|
|
|
|
Dallenne C, da Costa A, Decré D, Favier C, Arlet G (2010). Development of a set of multiplex PCR assays for the detection of genes encoding important β-lactamases in Enterobacteriaceae. Journal of Antimicrobial Chemotherapy 65(3):490-495.
Crossref
|
|
|
|
|
Eibach D, Dekker D, Gyau K, Wiafe C, Sarpong N, Belmar C (2018). Extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae in local and imported poultry meat in Ghana. Veterinary Microbiology 217:7-12.
Crossref
|
|
|
|
|
Feglo P, Adu SK (2016). Antimicrobial Resistance Patterns of Extended Spectrum Β -Lactamase Producing Klebsiellae and E. coli Isolates from a Tertiary Hospital in Ghana. European Scientific Journal 12(30):174-187.
Crossref
|
|
|
|
|
Ghazanfar A, Iahtasham K, Maskoor M, Sajjad-ur-Rahman, Tayyaba Y, Shahzad A (2019). High rates of CTX-M group-1 extended-spectrum β -lactamases producing Escherichia coli from pets and their owners in Faisalabad, Pakistan. Infection and Drug Resistance 12:571-578.
Crossref
|
|
|
|
|
Hackman HK, Brown CA,Twum-Danso K (2014). Antibiotic Resistance Profile of CTX-M-type Extended-Spectrum Beta-Lactamases in Escherichia coli and Klebsiella pneumoniae in Accra, Ghana. Journal of Natural Sciences Research 4(12):2225-2921.
|
|
|
|
|
Hartley SE, Valley SC (2014). Prevention of Catheter-associated urinary tract infections in the hospital. Hospital Medicine Clinic 4(2):258-271.
Crossref
|
|
|
|
|
Hooton TM, Bradley SF, Cardenas DD, Colgan R, Geerlings SE, Rice JC (2010). Diagnosis, Prevention, and Treatment of Catheter-Associated Urinary Tract Infection in Adults: 2009 International Clinical Practice Guidelines from the Infectious Diseases Society of America. Clinical Infectious Diseases 50(5):625-663.
Crossref
|
|
|
|
|
Hyun Y, Seung C, Jung I, Suck H, Ho C, Yu S (2015). Antimicrobial susceptibilities of extended‑spectrum beta‑lactamase‑producing Escherichia coli and Klebsiella pneumoniae in health care‑associated urinary tract infection: Focus on susceptibility to fosfomycin. International Urology and Nephrology 47(7):1059-1066.
Crossref
|
|
|
|
|
Kwadzo GT, Dadzie F, Osei-asare YB, Kuwornu JKM (2015). Consumer Preference for Broiler Meat in Ghana : A Conjoint Analysis Approach. International Journal of Marketing Studies 5(2):66-73.
Crossref
|
|
|
|
|
Köves B, Magyar A, Tenke P (2017). Spectrum and antibiotic resistance of catheter-associated urinary tract infections. GMS Infectious Diseases 5:1-5.
|
|
|
|
|
Lewis II JS, Herrera M, Wickes B, Patterson JE, Jorgensen JH (2007). First Report of the Emergence of CTX-M-Type Extended-Spectrum-Lactamases (ESBLs) as the Predominant ESBL Isolated in a US Health Care System. Antimicrobial Agents and Chemotherapy 51(11):4015-4021.
Crossref
|
|
|
|
|
Martin D, Fougnot S, Grobost F, Thibaut-Jovelin S, Ballereau F, Gueudet T, de Mouy D, Robert J (2016). Prevalence of extended-spectrum beta-lactamase producing Escherichia coli in community-onset urinary tract infections in France in 2013. Journal of Infection 72:201-206.
Crossref
|
|
|
|
|
Mensah DO, Nkrumah NO, Bonney EY, Mensah EO, Danso KT, Osei YD (2016). Genetic characterization of TEM-type ESBL-associated antibacterial resistance in Enterobacteriaceae in a tertiary hospital in Ghana. Annals of Clinical Microbiology and Antimicrobials, pp. 1-9.
|
|
|
|
|
Neupane S, Pant ND, Khatiwada S, Chaudhary R, Banjara MR (2016). Correlation between biofilm formation and resistance toward different commonly used antibiotics along with extended spectrum beta lactamase production in uropathogenic Escherichia coli isolated from the patients suspected of urinary tract infections visit. Antimicrobial Resistance Infectious Control 5(1):1-5.
Crossref
|
|
|
|
|
Nuhu T, Bolaji RO, Busayo O, Eugene OBB (2020). Prevalence of CTX-M-producing gram-negative uropathogens in Sokoto, North Western Nigeria. International Journal of Pharmacy and Pharmaceutical Sciences 12(1):21-25.
Crossref
|
|
|
|
|
Okoche D, Asiimwe BB, Katabazi FA, Kato L, Najjuka CF (2015). Prevalence and characterization of carbapenem-resistant enterobacteriaceae isolated from Mulago National Referral Hospital, Uganda. PLoS ONE 10(8):1-11.
Crossref
|
|
|
|
|
Parida S, Mishra SK (2013). Urinary tract infections in the critical care unit: A brief review. Indian Journal of Critical Care Medicine 17:370-374.
Crossref
|
|
|
|
|
Paterson DL, Ko W, Gottberg AVON, Casellas JM, Mulazimoglu L, Klugman KP (2001). Outcome of Cephalosporin Treatment for Serious Infections Due to Apparently Susceptible Organisms Producing Extended-Spectrum-Lactamases: Implications for the Clinical Microbiology Laboratory 39(6):2206-2212.
Crossref
|
|
|
|
|
Percival SL, Suleman L, Vuotto C, Donelli G, Percival SL (2018). Healthcare-associated infections, medical devices and biofilms: Risk, tolerance and control. Journal of Medical Microbiology 64:323-334.
Crossref
|
|
|
|
|
Reid R, Al-bayati M, Samarasinghe S (2018). Genotypic Identification of Extended-Spectrum β-Lactamase (ESBL)-Producing Enterobacteriaceae from Urinary Tract Infections in the Leicestershire Area, United Kingdom: A One Health Prospective. Journal of Infectious Disease Diagnosis 3(2):122:1000122.
|
|
|
|
|
Sana T, Rami K, Racha B, Fouad D, Marcel A, Hassan M (2011). Detection of genes TEM, OXA, SHV and CTX-M in 73 clinical isolates of Escherichia coli producers of extended spectrum beta-lactamases and determination of their susceptibility to antibiotics. International Arabic Journal of Antimicrobial Agents 1(1):1-6.
|
|
|
|
|
Shaikh S, Fatima J, Shakil S (2015). Antibiotic resistance and extended spectrum beta-lactamases: Types, epidemiology and treatment. Saudi Journal of Biological Science 22(1):90-101.
Crossref
|
|
|
|
|
Shanmugam K, Thyagarajan R, Katragadda R, Vajravelu L, Lakshmy A (2017). Biofilm formation and Extended Spectrum Beta Lactamases (ESBL) producers among the gram negative bacteria causing urinary tract infections. International Journal of Medical Microbiology and Tropical Diseases 3(8):86-90.
|
|
|
|
|
Subramanian P (2012). Antiobiotic resistance pattern of biofilm forming uropathogens isolated from catheterised patients in Pondicherry, India. Australasian Medical Journal 5(7):344-348.
Crossref
|
|
|
|
|
Subramanian P, Umadevi S, Kumar S, Stephen S (2012). Determination of correlation between biofilm and extended spectrum β lactamases producers of Enterobacteriaceae. Scholar's Research Journal 2(1):2.
Crossref
|
|
|
|
|
Verderosa AD, Totsika M, Fairfull-Smith KE (2019). Bacterial Biofilm Eradication Agents: A Current Review. Frontiers in Chemistry 7(824):1-17
Crossref
|
|
|
|
|
Vuotto C, Longo F, Pascolini C, Donelli G, Balice MP, Libori MF (2017). Biofilm formation and antibiotic resistance in Klebsiella pneumoniae urinary strains. Journal of Applied Microbiolgy 123:1003-1018.
Crossref
|
|
|
|
|
Zeynudin A, Pritsch M, Schubert S, Messerer M, Liegl G, Hoelscher M (2018). Prevalence and antibiotic susceptibility pattern of CTX-M type extended-spectrum β-lactamases among clinical isolates of gram-negative bacilli in Jimma, Ethiopia. BMC Infectious Diseases 18(1):1-10.
Crossref
|
|