ABSTRACT
This study characterized Cucumber mosaic virus (CMV) infecting pepper in Southwestern Nigeria with the aim of providing up-to-date information on the taxonomic subgroup(s) of the parading strains in the study area. Fifty pepper leaf samples with one or more symptoms of mottling, mosaic, necrosis, leaf distortion and stunting were collected from eight commercial farms across Southwestern Nigeria and screened for CMV using antigen coated plate enzyme linked immunosorbent assay (ACP-ELISA). From the seropositive samples, four representative samples were selected, labelled PSWN1-4 before subjected to reverse transcription polymerase chain reaction (RT-PCR) and nucleic acid sequencing using a pair of primers specific for the amplification of RNA 3 genomic fragment of CMV. These were followed by multiple nucleotide sequence alignment and phylogenetic estimations of the isolates alongside selected CMV strains that were reported in previous studies using the molecular evolutionary genetics analysis (MEGA) software. The result of the ACP-ELISA test revealed that 14 (28%) of the samples collected were positive for CMV infection. The resulting nucleotide sequences from RT-PCR revealed 98% nucleotide homologies with several corresponding sequences of CMV strains from the Genbank. Further analysis of the PSWN nucleotide sequences through multiple nucleotide sequence alignment revealed the absence of the EcoR1 restriction site found only within the examined genomic portion of RNA 3 of subgroup II strains. These features defined the detected isolates as subgroup I strains. Phylogenetic analysis and estimations of genetic distances, conclusively distinguished the pepper-infecting CMV isolates from Southwestern Nigeria as members of subgroups IA and IB, which are the most virulent subgroups.
Key words: Cucumber mosaic virus (CMV), pepper, Southwestern Nigeria.
Abbreviation:
BLAST, Basic alignment search tool; CMV, cucumber mosaic virus; EDTA, ethylenediaminetetraacetic acid; ELISA, enzyme linked immunosorbent assay; PSV, peanut stunted virus; RT-PCR, reverse transcription polymerase chain reaction; TAV, tomato aspermy virus.
Cucumber mosaic virus (CMV) infects more than one thousand two hundred plant species worldwide causing viral epidemics and economic loss in several economically important crops including tomato (Palukaitis and Garcia-
Arenal, 2003a). The virus causes mosaic symptoms in cucumber and other cucurbits; mosaic, systemic necrosis and shoestring-shaped leaf in tomato; mosaic and ringspot in pepper; withering in spinach; mosaic and stunting in clover, lupins and lucerne. They also cause stunting in soybean; mosaic and infectious chlorosis in banana; and mosaic and dwarfing in many other species of dicotyledonous and monocotyledonous plants (Palukaitis, 2003).
CMV has a worldwide distribution (both in tropical and temperate climates) and noted to have the widest host range for any plant virus (Palukaitis and García-Arenal, 2003b), including more than one thousand two hundred species in over a hundred families of angiosperm (
Douine et al., 1979; Edwardson and Christie, 1991).
Phylogeny based on analysis of RNA 3 sequences indicates that subgroup I strains can be further subdivided into subgroups IA and IB, in which subgroup IA strains are more closely clustered together than subgroup IB strains. Subgroups IA and II appear to be monophyletic (
Roossinck et al., 1999).
The presence of the characteristic EcoR1 restriction site, 5' GAATTC 3', located at a particular region within RNA 3 gene fragment of the subgroup II strains, has also been used by researchers to differentiate members of the subgroup II strain from Subgroup I (Eni et al., 2013). The EcoR1 restriction site is found around nucleotide 1866-1871 of the RNA3 of subgroup II (Kayode, 2014). Subgroup I strains have been shown to be more virulent in crops than the subgroup II strains (Wahyuni et al., 1992; Zhang et al., 1994). Cross protection occurs between strains from all subgroups.
In view of the potential devastating impact of the different CMV subgroups on pepper production, this study was designed to molecularly identify and characterize the CMV isolates infecting pepper on selected commercial farms in major pepper producing areas of Southwest Nigeria. This research work reported the incidence and classification of the common strains of CMV in pepper within Southwestern Nigeria into subgroups. The knowledge of the taxonomic subgroup(s) of the isolated pepper-infecting CMV strains in farms provided an insight into the degree of virulence of the parading strains of CMV in the study area, since virulence activity and devastating impact of CMV strains has been reportedly linked to strain’s subgroup membership.
Sample collection
Fresh, young expanded leaves were collected from pepper plants showing symptoms of CMV diseases. The samples were preserved in air-tight McCartney bottles pre-loaded with silica gel (Sigma Aldrich) and covered with cotton wool. Fifty (50) samples were collected from eight commercial farms (Table 1) within the study area.
Serological indexing for virus detection in collected samples
Antigen coated plate enzyme-linked immunosorbent assay (ACP-ELISA) method was used for the detection of CMV in collected leaf samples as described by Hughes and Tarawali (1999). From each of the leaf sample, 100 mg were ground in 1 ml of antigen extraction/coating buffer (1:10 w/v). One hundred microlitres of the antigen ground was dispensed into each well of the ELISA plate. The plate was covered and incubated overnight at 4°C, then washed three times with PBS-Tween by flooding for 3 min each time. The plate was drained, tap dried and blocked with 200 µl per well of 3% (w/v) dried non-fat skimmed milk in PBS-Tween. The plate was covered, incubated at 37°C for 30 min, emptied and tap dried. 100 μl of polyclonal antibody was added to each well and diluted with conjugate buffer in the ratio 1:2000. The plate was covered and incubated at 37°C for 1 h, then washed three times with PBS-Tween by flooding for 3 min each time before tap drying. 100 μl of goat anti-rabbit alkaline phosphatase conjugate diluted in conjugate buffer was added into the each well. The plate was covered and incubated at 37°C for 1 h. The plate was finally washed three times with PBS-Tween by flooding for 3 min each time before tap dried. 100 μl of 0.5 mg/ml of p-nitrophenyl phosphate substrate in substrate buffer was added per well. The plate was placed in the multiscan ELISA plate reader provided with 405 nm filter and the reading was taken after 1 h and overnight. Samples with values exceeding twice the reading of the healthy control were considered CMV positive.
Nucleic acid extraction
Four seropositive samples with the highest ELISA readings were selected for molecular characterization of the detected CMV. The samples were labeled PSWN1, PSWN2, PSWN3 and PSWN4. The cetyltrimethylammonium bromide (CTAB) method was used for nucleic acid extraction as described by Abarshi et al. (2010) and Dellaporta et al. (1983). CTAB RNA extraction buffer pH 8.0 was made by mixing 2% molecular biology grade cetyltrimethyl ammonium bromide powder (Sigma H6269) (w/v), 100 mM Tris-HCl, 20 mM EDTA, 1.4 M NaCl, and 0.2% β-mercaptoethanol (v/v) (was added just before use).
Procedure for RNA extraction
One hundred milligrammes of tomato leaf were ground in 1000 µl of nucleic acid extraction buffer in a sterile mortar and pestle. The sap was poured into new sterile tube and vortex briefly before incubating in water bath at 60°C for 10 min. The plant sap was brought to room temperature and equal volume of the mixture containing phenol, chloroform and iso-amyl alcohol in the ratio 25:24:1 was added. Sap was vortexed, centrifuged at 12000 rpm for 10 min and 450 µl of the supernatant was pipetted into new sterile tube. One microlitre of 1 U/µl DNase enzyme (Promega USA) was added to degrade the DNA portion of the nucleic acid. Cold isopropanol 300 µl was added, mixed and sap incubated for 1 h at -20°C. The mixture was centrifuged at 12000 rpm for 10 min to sediment the nucleic acids. The supernatant was gently decanted to ensure the pellets were not disturbed. 500 µl of 70% ethanol was added to the pellets and centrifuge at 12000 rpm for 5 min. The ethanol was decanted and the RNA air dried at room temperature. RNA pellets were suspended in 50 µl TE buffer for further use and storage at -80°C.
Reverse transcription polymerase chain reaction (RT-PCR)
With the aim of amplifying the 3' end of the coat protein (CP) gene and C-terminal noncoding region of RNA3 of CMV, RT-PCR was carried out using the CMV specific primers, 5' GCC GTA AGC TGG ATG GAC AA 3' and 5' TAT GAT AAG AAG CTT GTT TCG CG 3' as described by Wylie et al. (1993). Primers were synthesized by Inqaba Biotechnological Industries Limited, Walker Street 525, Muckleneuk 0002, Pretoria, South Africa. The PCR mix used for cDNA synthesis and amplification in a one step reaction contains 2 µl of the template RNA, 1 µl of 10 pm reverse primer, 1 µl of 10 pm forward primer, 3 µl of 25 mM MgCl2, 10 µl of PCR buffer, 1 µl of 10 mM dNTP, 0.24 µl of reverse transcriptase (1 U/µl) and 0.24 µl of Taq polymerase (1 U/µl) (Promega USA). RT-PCR was accomplished with the amplification programmed for one cycle cDNA synthesis of 44°C for 30 min and 95°C for 5 min and 35 cycles of amplification with 45 s of denaturation at 95°C, 45 s of annealing at 54°C and 45 s of extension at 72°C followed by one cycle of final extension for 7 min at 72°C. The RT-PCR products were analyzed by 1.5% agarose gel electrophoresis.
Sequence analysis
The PCR amplicons were purified with the addition of 70% ethanol and centrifugation at 9000 rpm for 45 s before carrying out sequencing. ABI 3130xL Genetic Analyzer (Applied Biosystems, California, USA), available at the Bioscience Center of IITA (International Institute of Tropical Agriculture) Ibadan was used for the nucleotide sequencing. Sequence similarity search of the GenBank database was done using the NCBI basic alignment search tool (BLAST) program.
Phylogenetic and molecular evolutionary analysis
A multiple nucleotide sequence alignment was conducted alongside corresponding CMV sequences from the Genbank sequence database, which have already been used in other studies. With the aim of determining the CMV taxonomy subgroup(s) of the PSWN isolates, the nucleotide sequences obtained were aligned with selected sequences of CMV subgroups IA, IB and II strains using the CLUSTALW program (Larkin et al., 2007). Tomato aspermy virus (TAV) and Peanut stunt virus (PSV) were used as out-group reference members of the genus Cucumovirus for rooting the phylogenetic tree. Corresponding sequences of CMV from neighbouring countries of Benin and Cameroun, available on the GenBank database were also selected for evolutionary comparison with the Nigerian strains. The analysis involved a total of 14 selected corresponding sequences from Genbank and 4 sequences obtained from this study. Phylogenetic and evolutionary relationship analysis was done on Molecular Evolutionary Genetics Analysis (MEGA) version 7 software package (Tamura et al., 2011) and a tree was created using the neighbour-joining method (Saitou and Nei, 1987). The evolutionary distances were computed using the Maximum Composite Likelihood method (Tamura et al., 2004) and are in the units of the number of base substitutions per site. Codon positions included were 1st+2nd+3rd+Noncoding. The optimal tree with the sum of branch length = 1.10493691 is shown. All positions containing gaps and missing data were eliminated. There were a total of 427 nucleotide positions in the final dataset.
ELISA test result and reverse transcription polymerase chain reaction (RT-PCR)
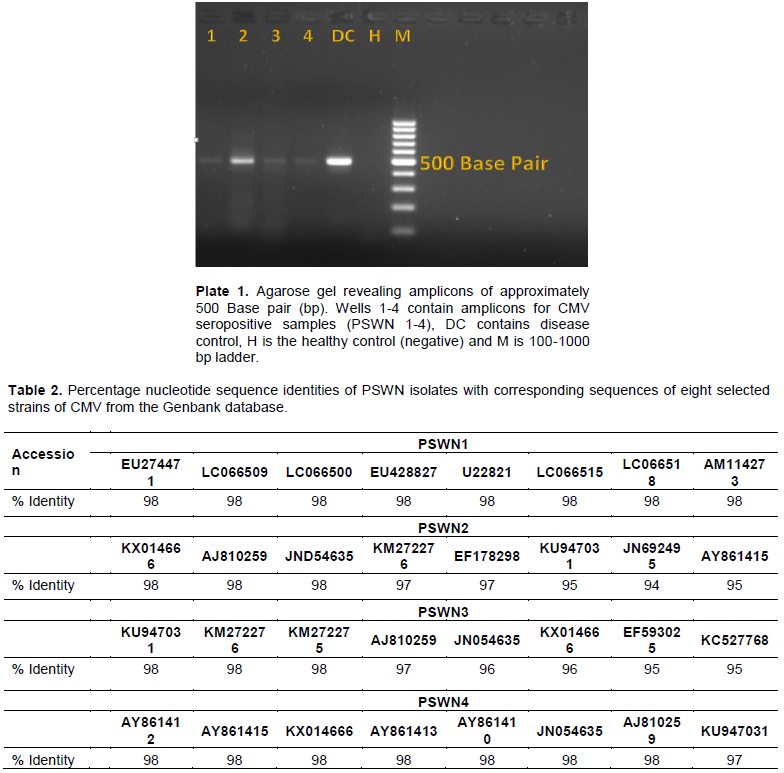
Fourteen (28%) of 50 samples showed the presence of CMV infection. All four representative samples selected for PCR and the disease control sample presented the expected amplicon bands of approximately 500 base pairs on the agarose gel (Plate 1). The positive control also revealed an amplicon of the same size as observed in the test samples. No amplicon was seen in the healthy control sample. This confirms the presence of CMV infecting pepper in Southwest Nigeria.
Sequence analysis and multiple nucleotide sequence alignment result
The obtained nucleotide (Appendix) sequences revealed high level similarities with corresponding CMV sequences available on the Genbank database. All four sequences recorded sequence identity of 98% with several CMV sequences in the Genbank. The percentage identities of the PSWN isolates with eight selected corresponding CMV sequences from the Genbank are shown in Table 2.
Multiple nucleotide sequence alignment revealed a very high sequence homology among the PSWN isolates. The alignment also revealed the existence of a higher degree of sequence homology between the PSWN isolates and subgroup I strains than exist between PSWN isolates and the subgroup II strains. The characteristic EcoR1 restriction site, 5' GAATTC 3', which was found only within nucleotide 338-343 of the subgroup II strains but absent in the subgroup I strains and PSWN isolates (as enclosed in the box in Figure 1), distinguished the PSWN isolates as members of the subgroup I. Tomato aspermy virus (TAV) and Peanut stunted virus (PSV) were defined as out-group members as both revealed a relatively low degree of homology with all the CMV strains.
Phylogenetic and molecular evolutionary analysis
The phylogenetic tree further confirmed that the CMV strains characterized in this study belong to subgroups IA and IB as the isolates clustered together with members of CMV subgroup I with >70% bootstrap support (Figure 2). Isolates PSWN 2 - 4 clustered into subgroup IA while PSWN 1 clustered into subgroup IB.
Previously reported CMV strains from Cameroon (accession EU 428827), Benin (accession EU274471) and Nigeria (accession KM091954) also clustered into subgroup IA. Hence, the detected PSWN isolates and reported CMV strains from the named African countries all belong to Subgroup I. Tomato aspermy virus and PSV strains remain the out-group members as they formed a distinct branch separated away from all the CMV strains.
The outcome of the ACP-ELISA diagnostic test adopted for detection of CMV in pepper leaf samples was effective. This implies that this method is still valid for surveillance study of CMV. The 3' end of the coat protein gene and C-terminal noncoding region of RNA 3 of the PSWN isolates that were amplified in this study produced amplicons of approximately 500 nucleotides as expected (Plate 1) (Wylie et al., 1993). Multiple nucleotide sequence alignment (Figure 1) of the obtained PSWN sequences (Appendix) alongside the corresponding sequences of selected CMV subgroups IA, IB and II strains using the CLUSTALW2 program (Larkin et al., 2007) revealed the presence of the EcoR1 site located at nucleotide 338-343 of the RNA 3 fragment of CMV subgroup II strains and its absence in CMV subgroups IA, IB and the PSWN isolates described by Eni et al. (2013). This clearly distinguished all the PSWN isolates as
members of subgroup I.
In addition, CMV strains from Benin (accession EU274471), the strain from Cameroun (accession EU428827) and strain from Nigeria (accession KM 091954) isolated previously from yam, banana and tomatoes, respectively were placed into subgroup I alongside the PSWN isolates in the phylogenetic tree (Figure 2). This is an indication that subgroup I stains of the virus is the most predominant subgroup in this region of Africa. These findings are supported by the report of Palukaitis and García-Arenal (2003b) that most CMV strains have a restricted geographic distribution.
The phylogenetic analysis and estimations of genetic distances in this study yielded results that correlated with of Roossinck et al. (1999) thus classifying the CMV strains into three distinct subgroups; IA, IB and II. The result of distinguished all the pepper-infecting CMV isolates from Southwestern Nigeria as members of subgroup I with three isolates (PSWN2, 3 and 4) classified as subgroup IA strains and PSWN1 as subgroup IB strains. The phylogenetic tree also revealed that CMV strains within each subgroup radiate from a single point of origin, indicating the evolution of each subgroup from a single common ancestry. Strains belonging to subgroups IA and II were observed to be more closely clustered within their respective group when compared with strains within subgroup IB. This indicates that a monophyletic relationship exist within the two subgroups while a polyphyletic relationship exist in subgroup IB as reported by Roossinck et al. (1999). These features validate the phylogenetic tree obtained, hence the veracity of the PSWN strains as subgroup I members is established.
The fact that CMV strains reported in this study and other crops in southwestern Nigeria and the neighbouring countries belong to subgroup I (Eni et al., 2013; Kayode et al., 2014), which is the virulent group, may be due to the fact that these areas are all located within the tropical region of Africa possessing identical climatic and vegetative features which are specifically conducive for the proliferation of the subgroups IA and IB strains (Kayode, 2018).
CMV has been detected in several plant seeds and its numerous host range includes cultivated crops and weed species. It is therefore possible that contaminated seed lots and alternative weed and crop host plants serve as sources of CMV inoculum to cultivated pepper in affected farms. High aphids population observed on many farmlands within the study area (Kayode, 2014, 2018) have been attributed to be responsible for the rapid spread of CMV from infected crops and weeds to cultivated pepper plants.
Although the occurrence of CMV has been reported in several crops including pepper in several countries worldwide, to our knowledge, this is the first molecular identification of CMV subgroups IA and IB in cultivated pepper in Southwestern Nigeria.
This information on the genetic diversity of the common CMV strains in the study area is of economic importance in the development of control methods. The awareness of the fact that Subgroup IA and IB strains of CMV which are the most virulent strains (Wahyuni et al., 1992; Zhang et al., 1994), isolated from pepper as reported in this study is an eye-opener to the need for urgent control and effective management measures to minimize crop losses within the study area.
Considering the fact that the presence of alternative host plants and availability of transmitting aphid populations are required for CMV outbreaks and epidemic in pepper, farmers are therefore advised to get rid of all perennial weeds and alternative host plants reservoir in and around proposed farm lands; leave a distance of at least 5 m between pepper farms and surrounding weeds; plant pepper earlier to avoid high aphid populations that usually occur later in the planting season; plant late settings as far as possible from fields used to produce early pepper and other vegetables; lookout for the first symptoms of any virus disease (if detected, farmers should pull out and burn the infected plants after spraying with insecticide to kill any insect vectors); early monitoring of aphid populations within the season and application of appropriate insecticide when necessary. Safety standards and regulations must be adopted during application of insecticides to prevent food poisoning and health hazards.
CMV as observed has been shown to present many symptom variants, making the identification often difficult from symptoms alone, the production of affordable, rapid, easy-to-use, on-the-spot ELISA diagnostic kit should be encouraged and facilitated to enhance surveillance studies, early detection and control of outbreaks and epidemics in the study area.
Other management strategies for the control of viral diseases of pepper include: planting of virus-free seeds obtained from credible sources rather than collection of seeds from previously harvested tomato fruits that might have been infected and raising of pepper seedlings in screen houses or nets using sterilized soil during the nursery stage.
BLAST, Basic alignment search tool; CMV, cucumber mosaic virus; EDTA, ethylenediaminetetraacetic acid; ELISA, enzyme linked immunosorbent assay; PSV, peanut stunted virus; RT-PCR, reverse transcription polymerase chain reaction; TAV, tomato aspermy virus.
The authors have not declared any conflict of interests.
REFERENCES
|
Abarshi MM, Mohammed IU, Wasswa P, Hillocks RJ, Holt J, Legg JP, Seal SE, Maruthi MN (2010). Optimization of diagnostic RT-PCR protocols and sampling procedures for the reliable and cost-effective detection of Cassava brown streak virus. Journals of Virological Methods 163: 353-359.
Crossref
|
|
|
|
Dellaporta SL, Wood J, Hicks JB (1983). A plant DNA minipreparation: version II. Plant Molecular Biology Reporter 1:19-21.
Crossref
|
|
|
|
|
Devergne JC, CardinL (1973). Serological relationship between Cucumovirus (CMV, TAV, PSV). Annals of Phytopathology 7: 255- 276.
|
|
|
|
|
Douine L, Quiot JB, Marchoux G, Archange P (1979). Census of plant species susceptible to Cucumber mosaic virus, Study bibliography. Annals of Phytopathology 11: 439-475.
|
|
|
|
|
Edwards MC, Gonsalve D (1983). Grouping of seven biologically defined isolates of Cucumber mosaic virus by mapping. Phytopathology 73: 1117-1120.
Crossref
|
|
|
|
|
Edwardson JR, Christie RG (1991). CRC Handbook of viruses infecting legumes, Boca Raton, CRC Press, 293.
|
|
|
|
|
Eni OA, Kumar PL, Aseidu R, Alabi OJ, Naidu RA, Hughes J d' A, Rey MEC (2013). Characterization of Cucumber mosaic virus isolated from yam (Dioscorea spp.) in West Africa. African Journal of Biotechnology 12(22):3472-3480.
|
|
|
|
|
Gonda TJ, Symons RH (1978). The use of hybridization analysis with complementary DNA to determine the RNA sequence homology between strains of plant viruses: Its application to several strains of cucumoviruses. Virology 88: 361-370.
Crossref
|
|
|
|
|
Hughes J d' A, Tarawali SA (1999). Viruses of herbaceous legumes in moist savanna of West Africa. Tropical Science 39:70-76.
|
|
|
|
|
Kayode AB (2014). Molecular identification and distribution of Cucumber mosaic virus infecting tomatoes in selected farms in southwestern Nigeria. M. Sc. Thesis, Obafemi Awolowo University, Ile-Ife, Osun State, Nigeria: 19
|
|
|
|
|
Kayode AB, Odu BO, Ako-Nai KA, Alabi OJ (2014). Occurrence of Cucumber mosaic virus subgroups IA and IB Isolates in tomatoes in Nigeria. Plant Disease 98(12): 1750.
Crossref
|
|
|
|
|
Kayode AB (2018). Determination of the occurrence and molecular characterization of major viruses infecting tomato in southwestern Nigeria. Ph. D. Thesis, Obafemi Awolowo University, Ile-Ife, Osun State, Nigeria: 127.
|
|
|
|
|
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, GibsonTJ, HigginsDG (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23(21): 2947-2948.
Crossref
|
|
|
|
|
Owen J, Palukaitis P (1988). Characterization of Cucumber mosaic virus. I. Molecular heterogeneity mapping of RNA 3 in eight CMV strains. Virology 166(2): 495-502.
Crossref
|
|
|
|
|
Palukaitis P (2003). Scottish Crop Research Institute, Invergowrie, Dundee DD2 5DA, UK.
|
|
|
|
|
Palukaitis P, Garcia-Arenal F, (2003a). Cucumoviruses. Advances in Virus Research 62: 241-323.
Crossref
|
|
|
|
|
Palukaitis P, García-Arenal F (2003b). Description of Plant Virus: Cucumber mosaic virus. No. 400 (Revised version of DPV 213).
View
|
|
|
|
|
Piazzolla P, Diaz-Ruiz JR, Kaper JM (1979). Nucleic acid homologies of eighteen Cucumber mosaic virus isolates determined by competition hybridization. Journal of General Virology 45: 361-369.
Crossref
|
|
|
|
|
Roossinck MJ, Zhang L, Hellward K (1999). Rearrangements in the 5' nontranslated region and phylogenetic analyses of Cucumber mosaic virus RNA 3 indicate radial evolution of three subgroups. Journal of Virology 76: 6752-6758.
|
|
|
|
|
Saitou N, Nei M (1987). The neighbor-joining method: A new method for reconstructing phylogenetic trees. Molecular Biology and Evolution 4:406-425.
|
|
|
|
|
Tamura K, Nei M, Kumar S (2004). Prospects for inferring very large phylogenies by using the neighbor-joining method. Proceedings of the National Academy of Sciences (USA) 101:11030-11035.
Crossref
|
|
|
|
|
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011). MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution 28: 2731-2739.
Crossref
|
|
|
|
|
Wahyuni WS, Dietzen RG, Handa K, Francki RIB (1992). Serological and biological variation between and within subgroup I and II strains of Cucumber mosaic virus. Plant Pathology 41: 282-297.
Crossref
|
|
|
|
|
Wylie S, Wilson CR, Jones RAC, Jones MGK (1993). A polymerase chain reaction assay for Cucumber mosaic virus in lupin seeds. Australian Journal of Agricultural Research 44: 41-51.
Crossref
|
|
|
|
|
Zhang L, Hananda K, Palukaitis P (1994). Mapping local and systemic symptom determinants of Cucumber mosaic cucumovirus in tobacco. Journal of General Virology 75: 3185-3191.
Crossref
|
|