Methicillin resistant Staphylococcus aureus (MRSA) keeps emerging as a pathogen of significant public health concern. In this study, the occurrence and antibiogram of MRSA from fresh milk and milk products sold in Nasarawa State, north-central Nigeria, were determined. A total of 180 samples comprising of fresh milk, bulk milk, nono and kindirmo were collected and examined for the presence of antibiotic-resistant phenotypes of MRSA using standard microbiological procedures and molecular confirmation using species-specific 23S rDNA Polymerase chain reaction (PCR). Also, the presence of mecA gene was determined from confirmed MRSA strains using PCR. The cumulative occurrence of MRSA as obtained from this study was 5.00% (9/180); with 7.14, 6.06, 4.55 and 2.94% occurrence in bulk milk, nono, kindirmo and fresh milk respectively. With respect to the sampling locations, 2.86, 6.89, 7.14, 3.57, 6.45 and 3.45% occurrence of MRSA were recorded from Nasarawa, Keffi, Akwanga, Wamba, Lafia and Doma local government areas respectively. There was no significant difference (P>0.05) in the occurrence of MRSA in both the sample types and sampling locations. All the 9 MRSA strains examined showed 100% resistance to ampicillin and amoxicillin/clavulanic acid. This study recorded 5 antibiotic resistance patterns among theMRSA strains with multiple antibiotic resistance (MAR) index of 0.3 and above. Only 3 of the 9 MRSA strains isolated harboured mecA gene. This study provides baseline data on the public health risks associated with the consumption of milk products sold in Nasarawa State, Nigeria.
Staphylococcus aureus has been implicated in many diseases of animals and humans, and the pathogenicity of this bacterium is mainly due to a combination of genetic factors mediating virulence, invasive ability, ability to evade the immune system of the host, and antibiotic resistance (Chua et al., 2014). Milk and milk products, especially those produced from raw milk under poor hygienic conditions serve as potential vehicles for the transmission of foodborne pathogens including antibiotic-resistant strains of S. aureus (Kadariyaet al., 2014).
Methicillin-resistant S. aureus (MRSA), including the livestock-associated methicillin-resistant S. aureus (LA-MRSA), has been isolated from dairy products such as raw milk and cheese globally (Peton and Le Loir, 2014). Nono and kindirmo are locally fermented milk products which are commonly produced and sold by Fulani pastoralists in Nigeria, especially in the northern part of the country. Since these products are produced under doubtful hygienic conditions, they could serve as vehicles for the transmission of MRSA.
In recent years, the spread of antibiotic-resistant bacterial pathogens, particularly multi-drug resistant zoonotic pathogens have raised increasing global concern (Ikeagwu et al., 2008). Related to this concern is the fact that, the use of antimicrobial agents in both human and veterinary medicine is promoting the steady rise in the prevalence of these antibiotic-resistant pathogens globally (Umaru et al., 2013). These antibiotic-resistant strains can be shed into the environment and consequently contaminate milk and milk products (Lee, 2003). MRSA harboursmecA gene (located on the staphylococcal cassette chromosome mec (SCCmec)) which codes for the production of an altered penicillin-binding protein (PBP2a or PBP2’) instead of the normal PBP that has a low affinity for beta-lactam antibiotics (Kwon et al., 2006; Scott and Van Duijkeren, 2010). The polymerase chain reaction (PCR) has high sensitivity and specificity in detecting methicillin resistance in S. aureus and is not affected by culture conditions such as temperature, pH, and the sodium chloride (NaCl) content of the growth medium which make the phenotypic determination of methicillin resistance in S. aureus vague (Zamani et al., 2007).Data on the occurrence and antibiotic susceptibility of MRSA in Nasarawa State, north-central Nigeria is scarce. Consequently, this study sought to determine the occurrence, antibiotic resistance patterns, and detection ofmecA by PCR technique in MRSA isolated from fresh milk and milk products in Nasarawa State, north-central Nigeria.
Study area
This study was conducted in Nasarawa State, North-CentralNigeria. Nasarawa State has with 13 Local Government Areas (LGA) and is situated between latitude 70° 40’ 0”N and 90° 40’ 0”N, and longitude 70° 0’ 0”E and 90°30’ 0”E. It isbounded by Taraba State to the southeast, Kaduna State to the north, Plateau State to the northeast, Kogi State to the southwest, and to the south and west by Benue State and the Federal Capital Territory (FCT), Abuja.
Sample size determination and sample collection
In this study, previously reported S. aureus prevalence of 12.60% (Umaru et al., 2013) was used for the determination of the sample size in the equation described by Naing et al. (2006). A total of 180 samples of fresh cow milk (34) and milk products consisting of bulk milk (14), locally fermented milk, nono(66) and locally-pasteurized milk, kindirmo(66) were randomly collected from each of the selected LGA including Akwanga and Wamba (Nasarawa North),Keffi and Nasarawa (Nasarawa West), and finally Lafia and Doma (Nasarawa South) respectively between May to October, 2018. Each of the collected sample was separately placed in sterile plastic bags, stored in cool box with ice and immediately transported to the microbiology laboratory of the Ahmadu Bello University, Zaria, Nigeria for microbiological analyses.
Isolation and Identification of S. aureus
A loopful of pre-enriched broth of each sample (25ml) diluted in 225ml of 1% buffered peptone water, homogenized and incubated for 18 h, and then inoculated onto the surface of prepared Baird-Parker agar (BPA) plates (Oxoid, Basingstoke, England) supplemented with 5% egg yolk tellurite emulsion (Baird-Parker, 1962). After 24 h incubation at 37°C,discrete colonies were selected based on theircultural and morphological features,and then sub-cultured onto selective media and nutrient agar for identification via biochemical tests (Patrick et al., 2013). S. aureus ATCC 25923 strain was used as a positive control.
Biochemical tests
The conventional biochemical tests carried out to identify the suspected S. aureuscolonieswereCatalase test, haemolysis on blood agar, slide coagulase test, and DNase test (Japoni et al., 2004).Also, the Microgen® Staphylococci Identification (STAPHidentification) kit was used for the identification of S. aureus following the manufacturer’ instruction.
Molecular characterization of the S. aureusstrains
The genomic DNA was extracted by boiling method as previously described by Akanbi et al. (2017). The concentration and the purity of extracted DNA from each strain was determined (Perez-Roth et al., 2001) and then used as DNA templates for the PCR.PCR reaction was performed using species-specific amplification of 23S rDNA (1250bp). The reaction was performed according to the method described by Shuiepet al. (2009). The forward and reverse primers used and PCR conditions were: 'F- ACGGAGTTACAAAAGGACGAC' and 'R –AGCTCAGCCTTAACGAGTAC' (Shuiep et al., 2009) (Inqaba, South Africa). The reaction mixture (25 µl) consisted of 1.0µl of each primer (forward and reverse) (10pmol/µl), 0.8µl dNTP (10mMol), 3.0 µl of 10x Taq buffer (Promega®, USA), with a final concentration of 1.8 µl MgCl2, (Promega®, USA), 0.1 µliTaqpolymerase (5U/µl) (Promega®, USA), 2.5 μl of genomic DNA, and finalivolumeiwasiadjusteditoi25iμlibyiaddingitheiremaining Volume iof inucleasei free-water. Amplification iwas icarriedi out withithermocycler (GeneiAMP®iPCRiSystem i9700, iABi Applied Biosystems,iSingapore) withianiinitialidenaturationiat i94°Cifor 5imin,ifollowedibyi37icyclesiofidenaturationiatii94°Cifor 40s, annealing temperature of primerswas 64°Cfor i60 s, extension at 72°C for75 sand the final extension was done at 72°C for 7min. A total of 10µl of the amplified PCR products werere solved by electrophoresis in 1.5% agarose gel (Amresco Bioscience, USA) with Trisacetate electrophoresis buffer(TAE, i4.0 mmol/Li Tris i1immol/LiEDTA, pH i8.0)at 100 V fori30imin, stainediwithiethidiumibromideiandifinallyivisualizediandidocumentedunderiUVitransilluminator(BIO-RAD Molecular Imager® Gel DocTM, Canada). The image was stored in a computer for further analysis.
Antibiogram of S. aureusstrains
The Kirby-Bauer agar disc diffusion method as described by the Clinical Laboratory Standards Institute (CLSI) (CLSI, 2013) was used for the determination of the antibiogram of the S. aureus strains. S. aureus strains were examined against eleven antibiotics with the following concentrations: amoxicillin/clavulanic acid (30µg), ampicillin (10 µg), gentamicin (30 µg), chloramphenicol (30µg), erythromycin (15 µg), imipenem (10µg), ciprofloxacin (5µg), cefoxitin (30 µg), sulphamethoxazole/trimethoprim (25µg), vancomycin (30 µg), and tetracycline (30µg) (Oxoid, England). Based on the diameter zone of growth inhibition, the results were expressed as sensitive, intermediate, or resistant using CLSI breakpoint (CLSI, 2013). S. aureus ATCC i25932 was used as the quality control standard strain.
Determination of multiple antibiotics resistance (MAR) Index of the S. aureusstrains
The MAR index was determined for each of the strain examined using the formula:
MARI= ix/y,
Where ‘x’ is the number of antibiotics to which the strain display resistance, and ‘y’ is the total number of antibiotics to which the test strain had been evaluated for sensitivity (Tula et al., 2013).
Molecular detection of mecAgene in the MRSA phenotypes
The genomic DNA was extracted and the concentration and purity determined as earlier described. PCR reaction was performed to detect the presence ofmecA in strains that phenotypically exhibited resistance to cefoxitin (methicillin) according to the method described by Strommenger etal. (2003).The presence of mecA gene was detected in the methicillin-resistant isolates using the forward and reverse primers: 'F-AAAATCGATGGTAAAGGTTGGC' and' R-AGTTCTGCAGTACCGGATTTGC' i(Inqaba, South Africa) that are expected to yield a PCR product of approximately 533 bp (Strommenger et al., 2003). The PCR was carried out in a 50 µl of reaction mixture with a PCR buffer containing 200 µm concentration of each deoxynucleosidetriphosphate (dNTP), 10imM Tris-Hcl(pH i8.3), 50 mM KCl, 2.5 mM MgCl2, 1.25 unit of Taqpolymerase (Promega,iGermany), 0.25iµM concentration of each primerand 2.5 µl of DNA template. Amplification was carried out in a thermocycler(GeneAMP®iPCRSystem i9700, ABApplied Biosystems, Singapore). DNAiamplificationiwasicarriedioutifor i50icyclesiin i50iμliofireaction mixtureiasifollows:Idenaturationiat i95°Cifor i30 is,I annealingiat i55°Cifor i30 is,I andiextensioniat i72°Cifori1imin,iwithiaifinaliextensioniati72°Cifori5imin.iPCRiproducts (10iμl)ibwerei analysediin i1.5%i agarosei gel.i Thei gelicontainingiamplifiediDNAiwasistainediwithiethidiumibromideisolution i(10img/ml)ifor i10iminiandidestainediwithidistillediwaterifor i10imin.iThei533ibpiPCRiproductsiwereivisualized and documented under UVtransilluminator (BIO-RAD Molecular Imager®GeliDocTM,Canada). DNA from a known mecA-carrying S.aureus ATCC33591 strain was used as PCR positive control.
Statistical analyses
The data obtained from this study were analyzed using the chi-square test which was used to determine the statistically significant difference between the occurrences of MRSA in the different sample types and sampling areas. Comparisons were considered statistically significant when P-values <0.05.
The cumulative occurrence of MRSA as obtained from this study was 5.00% (9). From the samples examined, 4 (6.06%), 1 (7.14%), 3 (4.55%) and 1 (2.94%) from nono, bulk milk, kindirmo and fresh milk were respectively contaminated with MRSA (Table 1) with respect to sampling location, Nasarawa, Keffi, Akwanga, Wamba, Lafia, and Doma local government areas were found to have 2.86, 6.89, 7.14, 3.57, 6.45 and 3.45% occurrence of MRSA from all the samples examined (Table 2). The results of antibiotic susceptibility pattern of the 9 MRSA strains examined using 11 antibiotics are presented in Table 3. The results revealed that 6 (66.7%) of the isolates were susceptible to sulphamethoxazole /trimethoprim, 8 (88.9%) were susceptible to vancomycin, 7 (77.8%) were susceptible to chloramphenicol, 4 (44.4%) were susceptible to erythromycin, 9 (100%) were susceptible to ciprofloxacin and gentamicin, 8 (88.9%) were susceptible to imipenem and 3 (33.3%) were susceptible to tetracycline respectively (Table 3). However, whereas all the MRSA isolates were resistant to ampicillin and amoxicillin/clavulanic acid, 4 (44.4%) were resistant to tetracycline while 2 (22.2%) were resistant to both sulphamethoxazole/trimethoprim and erythromycin. The results obtained further showed that imipenem, gentamicin, ciprofloxacin, chloramphenicol, vancomycin, and sulphamethoxazole/trimethoprim were the antibiotics to which the isolates examined were most susceptible.
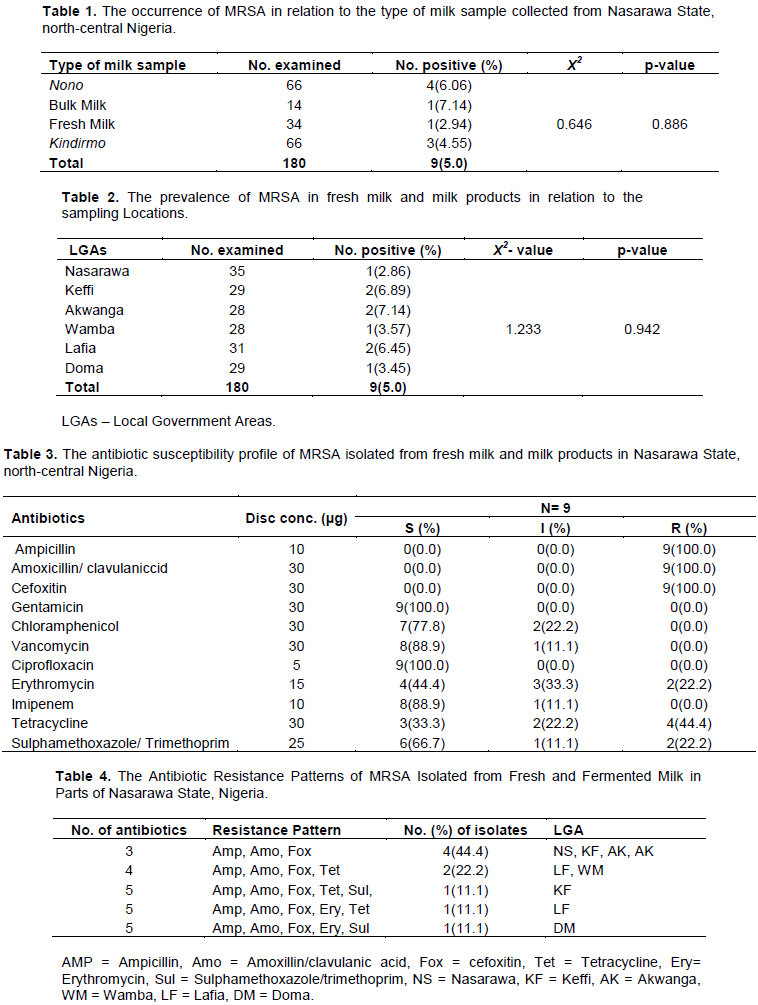
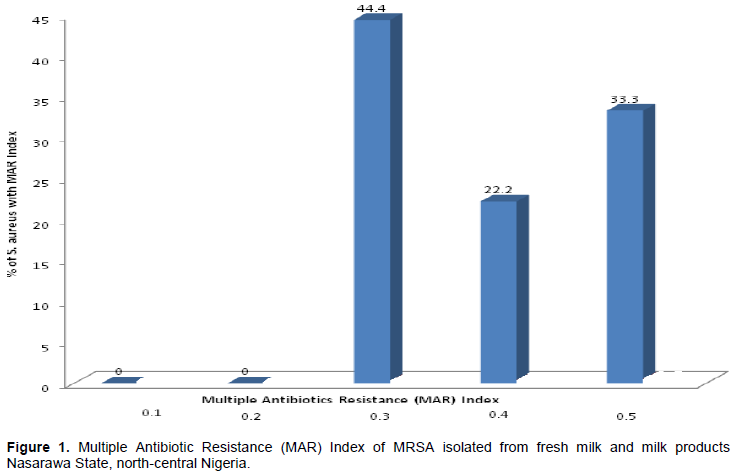
The results of the antibiotic resistance patterns of the 9 MRSA isolates obtained from fresh milk and milk products in Nasarawa State, north-central Nigeria is as presented in Table 4. A total of 5 antibiotics resistance phenotypes were obtained with varying multiple resistance types including 3, 4 and 5 different antibiotics combinations. All the isolates examined were resistant to 3 or more antibiotics. The highest frequency of multi-drug resistance patterns (as seen in three different isolates) were found to be made of 5 different antibiotics. Furthermore, the MAR indexes of the MRSA isolates obtained from this study are 0.3 and above (Figure 1). The results of the amplification of the species-specific 23S rDNA of the presumptive S. aureus isolates was molecularly confirmed with bands showing the amplicon size of 1250 bp positive for S. aureus (Figure 2). Furthermore, the presence of mecA gene was detected in only 3 of the 9 MRSA isolates examined by PCR. The 3 MRSA isolates withmecA-positive as evidenced by the amplification of themecA gene specific amplicon size of 533bp were: AKN1, NSN5, KFK12 (Figure 2).
Over the years, milk and milk products have been known as vehicles for the transmission of bacterial pathogens to man (Revathi et al., 2012). The contamination of food with antibiotic-resistant pathogens poses a significant global public health threat. The determinants of antibiotic resistance in these pathogens can be transferred to other bacteria of clinical significance. The cumulative occurrence of MRSA in fresh milk and milk products in Nasarawa State, north-central Nigeria as recorded in this study was 5.0%. However, higher MRSA occurrence of 12.14 and 12.60% from fresh milk and milk products in Kaduna and Zaria (which are major cities in north-western Nigeria) were previously reported (Umaru et al., 2013; Usman and Mustapha, 2016) respectively. Similarly, our result was also lower than the 8.70% prevalence of MRSA recorded from fresh and fermented milk in parts of Kaduna State, Nigeria (Okpo et al., 2016).
Furthermore, previous findings from other countries showed higher MRSA occurrence of 56, 55.26, 32 and 25.53% from dairy products in Turkey, Kenya, Algeria, and Bangladesh respectively (Patrickietial., i2013;I GundoganiandiAvci,i2014;iJahanietial., 2015; Chaalali eti al., i2016).iThe paucity of iinformation ioni MRSAi in milkiandimilkiproductsiinitheistudyiareaimadeiitidifficultitoimakeianyicomparisonianditoiassessitheileveliofiS.iaureusiinidairyiproductsiinitheiareasistudied. This study is the first comprehensive research conducted to carefully examine the occurrence of MRSA in dairy products in different municipal areas of Nasarawa State, north-central Nigeria, where these products are readily marketed and consumed.
The isolation of MRSA from milk products is a cause for public health concern because these products are regularly consume in the study area without considering the associated health hazards. The findings of this study also support the assertion that dairy products are one of the major vehicles for the transmission of MRSA to man. There was non statistically significant difference (P>0.05) recorded in the occurrence of MRSA among the fresh milk and milk products examined in this study. This can be attributed to the similar and uniform procedures employed during milking, processing, production and handling of these milk products within the study area.
Out of the nine MRSA strains isolated from the current study, one each was detected from fresh and bulk milk samples examined while four and three were isolated from nono and kindirmo samples respectively. This trend of MRSA occurrence is in contrast with the previous report of Umoh (1989) stating that fermented foods are generally not good media for the survival and growth of S. aureus. The occurrence of this organism in these locally processed foods may imply recontamination during and/or after processing. Notwithstanding, proper heat treatment and refrigeration can minimize the chances of contamination with S. aureus. It has also been observed that during the heat treatment of milk to produce kindirmo, the temperature does not rise enough to achieve effective pasteurization. The occurrence of S. aureus in fresh and bulk milk as recorded in this study may be attributed to the presence of subclinical mastitis in the milked cows poor sanitary practices during milking or use of unclean milking utensils. The main source of S. aureus in milk is the udder of infected cows which could be transferred via the milkers’ hands, milking utensils, towels, and the environment (Radostitsi et al., 1994). S. aureus can adapt and survive in the udder of cow and establish chronic and sub-clinical infections. From the udder, it can be shed into the milk which serves as a primary source of infection to individuals who consume unpasteurized milk.
The antibiotic susceptibility profile revealed that strains of MRSA examined were highly susceptible to ciprofloxacin (100%), gentamicin (100%), vancomycin (88.9%), imipenem (88.9%), and chloramphenicol (77.8%). The high susceptibility of these isolates to ciprofloxacin, gentamicin, and chloramphenicol was in consonance with other findings (Okpo et al. , 2016; Rodrigues et al., 2017), who recorded high susceptibility Of MRSA isolated from dairy products to these three antibiotics in areas in Kaduna State, north-western Nigeria and in Brazil respectively. The ihigh iperformance iof theseiantibiotics icould bei attributed ito itheir ismallimolecularisizes, aifactorithatienhancesitheiri solubilityiinidiluents,ithusienhancingitheiripenetrationipowerithroughitheicelliwalliintoitheicytoplasmiofitheitargetiorganismiwhereitheyiexertitheireffects (Okpo et al., 2016). This also agrees with the assertion of Maillard (2002) who opined that the high efficacy of these antibiotics in MRSA strains may be attributed to their molecular sizes.
Similarly, this study recorded a high level of MRSA strains susceptibility (88.9%) to vancomycin, a glycopeptide. This finding is not surprising because vancomycin is rarely used in the treatment of diseases in livestock in the study area. Also, the administration of vancomycin as chemoprophylaxis and antibiotic growth promoter which could further lead to resistance among bacteria as a result of selective pressure is not a known practice within Nasarawa State, Nigeria. In consonance with the findings of the present study, Suleiman et al. (2012) and Rodrigues et al. (2017) opined that the non-use of vancomycin for routine chemoprophylaxis and therapy in an area could result in high vancomycin susceptibility of strains of S. aureus isolated from such locality. The findings from this studyfurther agrees with the report of Alianet al. (2012) in Iran who recorded 0.0% resistance to vancomycin among S. aureus isolated from dairy products. However, our finding differs starkly with the report of Usman and Mustapha (2016), and Umaru et al. (2013) who reported 66.7% and 42.6% vancomycin resistance in MRSA strains isolated from fresh milk and milk products in Nigeria. The disparity between our findings and theiaforementionedicould beiasiairesultiof contaminationiofitheimilkiwithivancomycin-resistanti S. iaureus iderived fromihuman sources.
All the 9 MRSA strains examined from this study were resistant to amoxicillin/clavulanic acid and ampicillin. This could be attributed to the use and misuse of these antibiotics in the study area. Thisifindingiisinot surprising because outside ithei hospital ienvironment, ipeopleihaveieasyiaccessitoivariousiantibioticsiatianyidrugistoreiwithoutianyiprescriptionifromiqualifiedipersonnel. The findings of Anueyiagu and Isiyaku (2015) who reported 100% resistance of S. aureus isolated from dairy products in Jos, Plateau State, Nigeria and also in Bangladesh (Jahan et al., 2015) agrees with the results obtained from this study. Moreover, β- lactam antibiotics including ampicillin and amoxicillin/clavulanic acid are commonly used as dry-cow treatment. This may also have contributed to the increasing resistance among S. aureus strains to these drugs due to selective pressure. S. aureus resistant to one β-lactam drug can further develop resistance to other β-lactams drugs because they have the same mechanism of activity.
The relatively high level to tetracycline resistance as observed in this study could be attributed to tetracycline being the most commonly available antibiotic that is used as a growth promoter and routine prophylaxis in livestock management in Nigeria (Olatoye, 2010). Tetracycline resistance is a cause for immense concern considering that it is a first-line drug in Nigeria. Also, individuals with cases of gastrointestinal infections in Nigeria and most developing countries readily purchase tetracycline over-the-counter for self-medication (Chigor et al., 2010). Furthermore, MRSA strains from dairy products have been reported to show high instances of 55.5 and 40% resistance to tetracycline in Nigeria (Usman and Mustapha, 2016) and Ethiopia (Tessema, 2016). This trend is a cause for concern in both human medicine and livestock production due to the existing emergence of bacterial strains resistant to major antibiotics.The use of antibiotics either as prophylaxis or growth promoters in food animals are now regarded as the major avenue for the spread of resistant bacteria through the food chain to humans result in severe human infections (Phillips et al., 2004).
The relatively high resistance exhibited by MRSA strains to erythromycin in the study area reflects its frequent use and abuse. Higher levels of 85.7 and 76% resistance among S. aureus isolated from dairy products have been reported in Nigeria (Anueyiagu and Isiyaku, 2015)and Iran (Mirzaei et al., 2012) respectively. Also, the relatively high levels of resistance to sulphamethoxazole/trimethoprim as recorded in this study is baffling, considering the fact that the drug is not routinely used in veterinary practice in Nigeria. This may suggest cross-contamination of the dairy products by handlers with the drug-resistant pathogens. Furthermore, mixed fermentation is known to occur in local milk products includingnono andkindirmo. As a result of such uncontrolled fermentation process, different organisms can be involved at different times, hence, transferring determinantsiofiantibioticiresistanceibetweenithem.
Foodiisianiimportantimedium through which ithe itransfer iof determinantsiofiantibioticiresistancei\ amongibacteria occur. Such transfer can occur by means of residues of antibiotics in foods, transmission of resistant pathogens, or by the ingestion of resistant foodborne pathogens (Pereira et al., 2009).
Multi-drug resistance is defined as the resistance of an isolate to two or more antibiotics (Olayinka et al., 2004). All the nine MRSA strains isolated from this study showed multidrug resistance to the antibiotics tested at different percentages. The isolates were resistant to a combination of three, four, and five antibiotics tested. Isolates obtained from Keffi, Akwanga, and Lafia showed higher frequencies of multi-drug resistance. The multi-
drug resistance recorded among all the MRSA strains examined in this study agrees with other findings (Umaru et al., 2013; Anueyiagu and Isiyaku, 2015; Chaalal et al., 2016; Tessema, 2016), who reported cases of multi-drug resistance among MRSA strains isolated from dairy products in Zaria and Jos (Nigeria), Ethiopia, and Algeria respectively. Multi-drugiresistanceiiniS.iaureusimayibe attributed iin ipart,I toi the spread iof mobile geneticielementsilikeiplasmids,itransposons,iandiintegronsithatimayiconferiresistanceitoinumerousiantimicrobialiagents i(Zhaoietial., i2001). According itoiAarestrup(1995),I theideterminantsiofimulti-drugiresistanceiarecapablei ofi beingidisseminatediiniairegionioribetweeniregionsiasiairesultiofiantibioticiselectiveipressureiinieitherilivestockiorihumans.iEmpiricalievidenceiaboundsiwhichiindicateithatidru g-resistantistrainsiofibacteriaicanibeitransmitteditoihumansiviaifood (Khachatourians, 1998; Revathi et al., 2012; Tessema, 2016).
This study recorded 5 antibiotic resistance patterns among the MRSA strains examined which varies from the 25 antibiotic resistance patterns recorded among S. aureus strains isolated from dairy products in Bahir Dar, Ethiopia (Shiferaw and Ahmad, 2016). The disparity in the antibiotic resistance patterns of MRSA strains recorded in the present study and the previous results recorded in Ethiopia could be as a result of the different levels of abuse of antibiotics in the two areas. All the MRSA strains examined in this study had a MAR index of 0.3. MAR index gives an indirect indication of the probable source of an organism.An organism originates from an environment with antibiotics abuse when its MAR index is greater than 0.2(Furtula et al., 2013).
Results from this study revealed ciprofloxacin, gentamicin, vancomycin, imipenem, and chloramphenicol as drugs of choice for MRSA infections in the study area since majority of the isolates were susceptible to them. The public health significance of the present findings is that antibiotic-resistant strains of S. aureusfrom milk products (or dairy animals) may be transmitted from the food chain to humans through occupational exposure, contact or wastewater run-off fromproduction sites. More so, in Nigeria, the indiscriminate use of antibiotics in livestock production could have alsocontributed immensely to the patterns of antibiotic resistance recorded in this study.
The detection ofmecA gene is the gold standard for the determination of MRSA from either clinical, food or environmental samples. Three (AKN1, NSN5, and AKF2) out of the nine MRSA strains examined harbourmecA gene with an approximate amplified PCR product size of 533 bp, indicating that the phenotypic resistance exhibited by the strains to cefoxitin (methicillin) was due to the possession ofmecA gene. This finding concord with the report of Umaru et al. (2013), who detected mecA gene in only four out of the 18 MRSA strains isolated from raw milk and milk products in Nigeria. Our findings also agree with the results obtained by Suleiman et al. (2012) who also reported the presence ofmecA gene in only 2 out of the 26 MRSA strains isolated from milk in Plateau State, Nigeria. Similarly, out of the four MRSA strains isolated from milk in Brazil, Rodrigues et al. (2017) reported the presence of mecA gene in only two of the four MRSA strains examined.
Conversely, however, the finding of this study differs with the results obtained by Usman and Mustapha (2016) in which no mecA gene was detected in all of the nine MRSA strains isolated from skimmed milk and yogurt in Kaduna metropolis, Nigeria. In the same vein, mecA gene was not detected in all of the 15 MRSA strains isolated from raw milk in Malaysia (Shamila-Syuhada et al., 2016).
The disparity between the results obtained in this study and those of the other researchers mentioned above can be attributed to the assertion of Lee et al. (2004), who posited that phenotypic resistance of S. aureus to cefoxitin (methicillin) could vary depending on cellular growth conditions. Phenotypic expression of resistance to methicillin in MRSA varies, and each istrain ihas iai characteristiciprofile ofitheiproportioni of bacterial icellsithatigrowiatispecificiconcentrationsiof imethicillin (Plata et al., 2013). It has also been observed that PCR detection of mecA gene does not always give indisputable results (Adesida et al., 2005). Some MRSA strains have been found to bemecA-negative in PCR, but resistant to cefoxitin (methicillin). Furthermore, some MRSA strains have also been reported to bemecA-positive but susceptible to cefoxitin (methicillin) (Olonitola et al., 2007). The absence of mecA in MRSA strains could also be an indication of the potential presence of ‘modified S. aureus’ (MODSA). MODSA possess modified penicillin-binding proteins (PBPs) which is a different classical mechanism of resistance to methicillin in MRSA (Bhutia et al., 2012). In this study, a negative correlation was found between the phenotypic resistance to cefoxitin (methicillin) and PCR results for the detection ofmecA gene that code for resistance to methicillin.The antibiotic susceptibility profile of the MRSA strains isolated from fresh milk and milk products showed high levels of resistance to ampicillin, amoxicillin/clavulanic acid and tetracycline. This finding is of great public health concern because these antibiotics are commonly used in Nigeria either therapeutically in human and veterinary medicine or as growth promoters and prophylaxis in animal production.This study finally established the presence of mecA gene in only three out of the nine MRSA strains isolated from fresh milk and milk products in the study area. There was also an indication of a negative correlation between the phenotypic resistance to cefoxitin (methicillin) and the presence ofmecA gene which have been established to code for resistance to methicillin in MRSA strains.