ABSTRACT
Food borne illnesses have major social and economic impacts. Escherichia coli O157:H7 is associated with food borne illness in human beings. It has been an important food borne pathogen that causes food borne diseases such as diarrhea, hemolytic uremic syndrome and hemorrhagic colitis. This study was conducted to detect the presence of E. coli O157: H7 in different food samples sold in Nigerian local markets. A total of 60 different food samples (3 each of meat, fufu, waterleaf, pumpkin, carrot, tomatoes, meat pie, yoghurt, watermelon, cucumber, groundnut, cabbage, garden egg, bread, okra, apple, chicken, unpasteurized milk, salad and pawpaw) were collected randomly from different markets in Calabar, Nigeria. The samples were analyzed using standard microbiological techniques. Isolation was carried out using pour plate technique on sorbitol MacConkey agar. The isolates were identified by morphological and biochemical tests. Out of the 60 samples investigated, 36 (60%) were found to be contaminated with E. coli O157:H7 while 24 (40%) were negative by conventional methods. All the isolates obtained from the samples were subjected to various biochemical tests and were all confirmed to be E. coli O157:H7. The occurrence of E. coli O157:H7 serotype in these food products indicates that there may be a potential risk for public health from consuming these foods. This study clearly indicated the need for proper handling and processing of food products especially ready to eat food products. It is also important that at household level proper hygienic measures should be taken to avoid cross contamination.
Key words: Escherichia coli O157:H7, diarrhea, food borne illnesses, food samples, hemorrhagic colitis.
Escherichia coli are large and diverse group of bacteria. It is the type of the genus Escherichia that contained mostly motile Gram negative bacilli that fall within the family Enterobacteriaceae. It is the predominant facultative anaerobe of the human colonic flora. The organism typically colonizes the infant gastrointestinal tract within hours of life, and thereafter E. coli and the host derive mutual benefit for decades (Kaper et al., 2004). E. coli is a bacterium that normally lives in the intestines of human and animals, the growth and survival of E. coli depends
on the number of environmental factors such as temperature, pH, water activity, composition of the food, carriage by cattle and contamination of surface water (Center for Disease Control, 2001). The temperature range of growth of E. coli is 7 to 8 to 46°C, with an optimum temperature of 35 to 40°C. Although most serotypes of E. coli are harmless, several produce toxins that cause illness. Some strains of E. coli including E. coli O157:H7, produce toxins known as shiga toxins and are called shiga toxin producing E. coli (STEC). Their virulence characteristics suggest that they may have significant impact on public health (Perera et al., 2015).
During the past two decades, disease caused by E. coli O157:H7 has been increasing (Mean et al., 2015). Currently, the Centers for Disease Control and Prevention (CDC) estimated that E. coli O157:H7 caused an average of 500 outbreaks that affect >73,000 persons and result in >61 deaths each year in the United States (Charatan, 2014). The epidemiology of E. coli O157:H7 has become an important research topic as manure harboring E. coli O157:H7 is dispersed, and soil, food, and water are cross-contaminated with feces containing E. coli O157:H7 (CDC, 2001; Nakazawa and Akiba, 2001; Mean et al., 2015).
E. coli O157:H7 is an important emerging human pathogen causing haemmorhagic colitis (HC), haemolytic uraemic syndrome (HUS) and thrombotic thrombocytopaenic purpura (TTP) (Amani et al., 2015; Fan et al., 2019). E. coli O157:H7 serotypes are identified as enterohaemorrhagic E. coli (Oksuz et al., 2004). The infections by E. coli O157:H7 have been reported of increasing frequency from all parts of the world in the form of food poisoning outbreaks (Jo et al., 2004; CDC, 2018). Because of the severity of these illnesses and the apparent low infective dose (<10 cells) (Bach et al., 2002), E. coli O157:H7 is considered one of the most serious of known foodborne pathogens (Blanco et al., 2003).
Enterohemorrhagic E. coli O157:H7 is an important food borne pathogen a causative agent of HC and HUS. Globally, STEC caused 2,801,000 acute illnesses annually, with an incidence rate of 43.1 cases per 100,000 persons per year. This burden led to 3890 cases of HUS and 230 deaths (Lupindu, 2017). Large outbreaks of EHEC infection were reported throughout the world. E. coli O157:H7 is the most commonly recognized STEC in the United States, however, many other STEC serogroups including O26, O103, O111, and O145, have been associated with outbreaks and sporadic cases of HC and HUS worldwide (Essendoubi et al., 2019). One of the largest E. coli O157:H7 (one of the serotypes of EHEC) outbreaks associated with food consumption occurred in Sakai City, Japan in 1996. About a quarter of African countries have reported isolation of STEC 0157:H7 either from humans, animals, food or the environment (Lupindu, 2017; Yusuf et al., 2018).
The overall aim of this study was to investigate the presence of E. coli O157:H7 in different food samples sold in Calabar Metropolis, Nigeria and to determine its incidence rate.
Sample collection
A total of 60 different food samples (3 each of meat, fufu, waterleaf, pumpkin, carrot, tomatoes, meat pie, yoghurt, watermelon, cucumber, groundnut, cabbage, garden egg, bread, okra, apple, chicken, unpasteurized milk, salad and pawpaw) were collected randomly from different markets in Calabar, Nigeria. All the samples were collected aseptically in sterile universal containers and polyethylene bags and immediately placed in pre-cooled containers containing ice packs and then transported to the laboratory for analyses.
Preparation of samples
About 25 g of food samples was taken and homogenized with 225 ml of buffered peptone water 0.1% in a stomacher for 15 min, after which the homogenate was used for the isolation.
Isolation of E. coli 0157:H7
Isolation was carried out after pre-enrichment of the samples by selective plating as described by Kim et al. (2005). One milliliter of the homogenate was used for ten-fold serial dilution after which 0.1 ml of 103 dilution factor was inoculated on Sorbitol MacConkey agar (SMAC) supplemented with cefixime (0.05 mg/l) and potassium tellurite (2.5 mg/l) in triplicate. The plates were then incubated at 28°C for 18 to 24 h. After the incubation period, the plates were observed for the growth of E. coli 0157:H7 colonies.
Purification and maintenance of isolates
Each discrete colony on a Petri dish was transferred using a sterile inoculating loop into plates containing freshly prepared Nutrient agar (NA) and were incubated at 37°C for 24 to 48 h, respectively. The isolates were then preserved on NA slants stored in the refrigerator at 4°C.
Biochemical confirmation of isolates
The suspected colonies of E. coli 0157:H7 were subjected to various tests and confirmed based on the biochemical characteristics. The individual colonies of EHEC from CT-SMAC agar were transferred to tryptic soy broth (TSB) and incubated at 37°C for 24 h. Primary identification tests like Gram's staining, catalase test and oxidase test were performed. Secondary identification tests like indole production, methyl red (MR) reaction, voges proskauer (VP) reaction, citrate utilization test, urease activity, and carbohydrate utilization test were carried out as per the standard procedures.
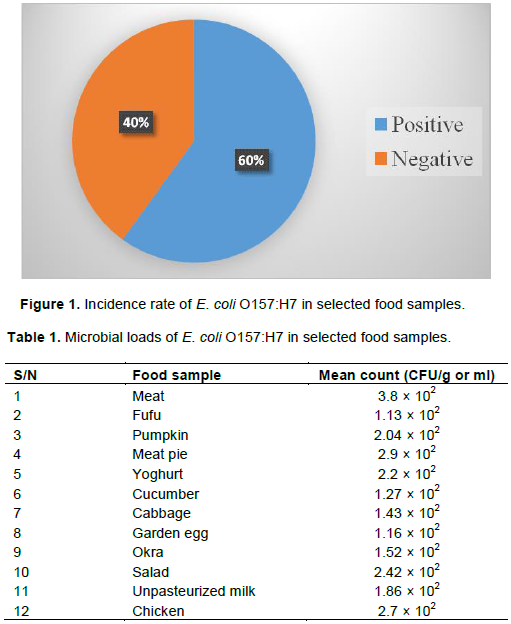
The percentage of E. coli O157:H7 in the food samples analyzed is shown in Figure 1 where the highest rate of occurrence of E. coli 0157:H7 was observed to be 60% with 40% representing negative occurrence. The results of the microbial load of E. coli O157:H7 in food samples analyzed are shown in Table 1. The highest E. coli O157:H7 load was recorded of meat sample with a mean load of 3.89 × 102 CFU/g while the least load was observed in fufu (1.13 × 102 CFU/g) sample.
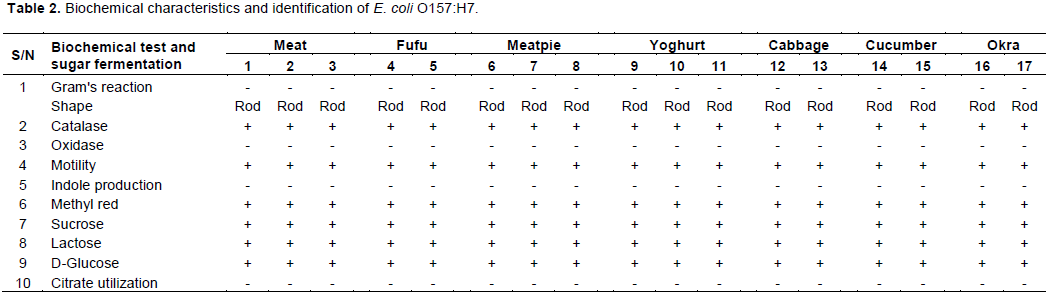
All these isolates were positive for catalase test, motility, indole production, MR, triple sugar iron agar reaction and Lysine decarboxylase. These isolates were negative for oxidase, VP, citrate utilization, urease and sodium chloride tolerance test. The results of these tests clearly indicated that they belonged to the category of enterohaemorragic E. coli. These isolates were then subjected to carbohydrate utilization tests for identification of species. The results suggested that all the 16 isolates were showing reaction similar to that of E.
coli O157:H7 shown in Table 2.
The results from the study revealed an overall incidence of E. coli O157:H7 as 60% (12/20) in all the collected food samples from different locations in Calabar, Nigeria. Kumar et al. (2004) found that 100% of different beef samples positive for E. coli O157:H7 in a study conducted in Mangalore, India. The United States Department of Agriculture (USDA, 2007) reported that the Food Safety and Inspection Service (FSIS) identified more than 75% of the ground beef and vegetable samples were positive for the presence of E. coli 0157:H7. Grant et al. (2011) reported that the prevalence of non 0157 EHEC in raw beef as 2.4 to 49.6% in Canada and United States it ranged from 5.7 to 26.2%. These suggested that foods, particularly beef are an important source of E. coli 0157:H7 infections. In the present study also, the highest mean load was observed in meat sample. This may be due to the fact that the gastro intestinal tract of the cattle is the most important predilection site of the organism (Vijayan et al., 2017). The range of food samples positive in this study for E. coli 0157:H7 is of concern to consumers and food processors. More of concern is the fresh vegetables that are eaten raw especially in salads and the already prepared foods.
The occurrence of E. coli O157:H7 of the present study was comparatively lower than a study done by Zahraa et al. (2016). They got a prevalence of 82% (135/164) from different food samples. Almost similar result was found in other studies conducted by Vinothkumar et al. (2014) in Puducherry, India who observed 84% prevalence in food samples. Momtaz and Jamshidi (2013) in Iran found 71.2% of the food samples were positive for E. coli 0157:H7. These findings suggested that cross-contamination of these food samples may occur in retail food shops and markets with higher prevalence in animals. Cattle act as a reservoir host for EHEC O157:H7 resulting in higher food contamination (Bindu and Krishnaiah, 2010). This is a serious health issue that the public health department of any government to take off. It is important that the regulatory agencies put up an enlightenment campaign to educate consumers on how to control or eliminate cross contamination by cooking meat properly, drinking pasteurize milk and juice, wash produce thoroughly, wash utensils very well, keep raw foods separate and to wash hands thoroughly after handling raw meats.
Similar study carried out in Iraq, from 100 samples of meat only two isolates and from 98 dairy product samples were detected as E. coli O157:H7 (Dhaher et al., 2010). In Iran, another study proved that, from 130 bulk tanks of milk just one isolate was E. coli 0157:H7 (Brenjchi et al., 2011). While, in another study, from 125 samples of soft cheese prepared from raw milk, found 5 isolates of E. coli 0157:H7 (Najand and Khalilli, 2007). Out of 50 ground beef samples, 7 strains of E. coli 0157:H7 were detected, while none was isolated from chicken drumsticks in Turkey (Fatma
and Murat, 2000).
In Turkey, studies conducted to detect E. coli O157 and/or E. coli O157:H7 revealed that E. coli 0157 have been isolated from different food products with an occurrence varying from 0 to 55% (Elmali et al., 2005). Few studies, however on the isolation of E. coli 0157:H7 from ground beef and vegetable products in Turkey have revealed negative results (Siriken and Pamuk, 2004; Siriken et al., 2004). Likewise, there is also no report of any outbreaks due to E. coli 0157:H7 in Turkey (Agaoglu et al., 2000). It is likely that these kinds of cases have not been reported or the causative agents of food poisonings have not been identified. Lack of direct link between isolates from humans and other sources makes it difficult to point out incident specific determinants and direction of transmission (Lupindu, 2017). But, it seems that incidences found in this study seem to be higher than those previous studies for other food products. Vegetables, milk and water have also been implicated in E. coli 0157:H7 poisoning outbreaks (Kayisoglu et al., 2003).
In Africa, a study carried out in Gwagwalada, Federal Capital Territory, Nigeria, among children between the ages of 0 to 24 months found 31.1% positive for STEC (Onanuga et al., 2014), which could be as a result of the mothers hygienic status.
The findings from this study suggested that most raw foods are contaminated with E. coli 0157:H7. This is a serious health issue. Cross-contamination of foods may be occurring in retail meat shops because studies have indicated higher prevalence of E. coli 0157:H7 in animals. Cattle act as a reservoir host for EHEC 0157:H7 resulting in higher food contamination. Mainly in the beef retail shops, beef carcasses were not hoisted but kept on the tables or the floor for dressing. More precautions are therefore needed processing and handling of meat to avoid cross contamination of foods. This study has also highlighted the contaminated levels of the various food products. The hygienic environments and handling could have contributed immensely to the cross contamination of other food products not prone to contamination by E. coli 0157:H7. Undocumented frequent food borne disease outbreaks in this part of the country could be attributed to E. coli 0157H:7.
Efforts should therefore be made to control this bacterium in Nigerian food products in order to avert any sickness or death resulting from eating food contaminated with E. coli 0157:H7.
The authors have not declared any conflict of interests.
REFERENCES
|
Agaoglu MT, Yavuz M, Berktas K, Guducuoglu H (2000). Detection of Escherichia coli 0157:H7 in retail ground beef, raw ground beef patties and raw meat balls sold in Van. Eastern Journal of Medicine 5:73-75.
|
|
|
|
Amani J, Ahmadpour A, Fooladi AAI, Nazarian S (2015). Detection of E. coli O157:H7 and Shigella dysenteriae toxins in clinical samples by PCR-ELISA. Brazilian Journal of Infectious Diseases 19(3):13-27.
Crossref
|
|
|
|
|
Bach SJ, McAllister TA, Veira DM, Gannon VPJ, Holley RA (2002). Transmission and control of Escherichia coli 0157:H7-A review. International Journal of Food Microbiology 95(1):41-49.
|
|
|
|
|
Bindu CK, Krishnaiah N (2010). Detection of Escherichia coli 0157:H7 prevalence in foods of animal origin by cultural methods and PCR technique. Veterinary World 3(1):13-16.
Crossref
|
|
|
|
|
Blanco JE, Mora, A, Rey J, Alonso JM, Hermoso M, Hermoso J, Alonso MP, Dahbi G, González EA, Bernárdez MI (2003). Serotypes, virulence genes and intimin types of shiga toxin (verotoxin)-producing Escherichia coli isolates from healthy sheep in Spain. Journal of Clinical Microbiology 41(4):1351-1356.
Crossref
|
|
|
|
|
Brenjchi M, Jamshidi A, Farzaneh N, Bassami MR (2011). Identification of shiga toxin producing Escherichia coli 0157:H7 in raw cow milk samples from dairy farms in Mashhad using multiplex PCR assay. Iranian Journal of Veterinary Research 12:27-35.
|
|
|
|
|
Center for Disease Control (CDC) (2001). Factors contributing to the emergence of E. coli 0157:H7 in Africa. Emerging Infectious Diseases 7(5):13-20.
|
|
|
|
|
Center for Disease Control (CDC) (2018). Thirty-one more sickened by E. coli-tainted romaine lettuce. Center for Infectious Disease 25(3):2-8.
|
|
|
|
|
Charatan F (2014). New York outbreak of E. coli poisoning affects 1000 and kills two. Brazilian Medical Journal 86:873-887.
Crossref
|
|
|
|
|
Dhaher FH, Mohammed DHA, Ali RM, Jamil MM (2010). Prevalence of E. coli 0157:H7 in beef meat products and dairy products sold in Baghdad local markets. Iraq Journal of Market Research and Consumer Protection 2:67-76.
|
|
|
|
|
Elmali M, Ulukanli Z, Tuzcu M, Yaman H, Cavli P (2005). Microbiological quality of beef doner kebabs in Turkey. Archiv fur Lebensmittelhygiene 56:32-34.
|
|
|
|
|
Essendoubi S, Stashko N, So I, Gensler G, Rolheiser D, Mainali C (2019). Prevalence of Shiga toxin-producing Escherichia coli (STEC) 0157:H7, Six non-0157 STECs, and Salmonella on beef carcasses in Provincially Licensed Abattoirs in Alberta, Canada. Food Control 105:226-232.
Crossref
|
|
|
|
|
Fan R, Shao K, Yang X, Bai X, Fu S, Sun H, Xu Y, Wang H, Li Q, Hu B , Zhang J, Xiong Y (2019). High prevalence of non-O157 Shiga toxin-producing Escherichia coli in beef cattle detected by combining four selective agars. BMC Microbiology 19:13-235
Crossref
|
|
|
|
|
Fatma B, Murat G (2000). The occurrence of Escherichia coli 0157:H7 in the ground beef and chicken drumsticks. Journal of Food Safety 2:13-15
|
|
|
|
|
Grant MA, Hedberge C, Johnson R, Harris J, Logue CM, Meng J (2011). The significance of non-0157 shiga toxin-producing Escherichia coli in food. USDA executive summary. Food Protection Trends 11:33-45.
|
|
|
|
|
Jo MY, Kim JH, Lim JH, Kang MY, Koh HB, Park YH, Yoon DY, Chae JS, Eo SK, Lee JH (2004). Prevalence and characteristics of Escherichia coli 0157 from major food animals in Korea. International Journal of Food Microbiology 95(1):41-49.
Crossref
|
|
|
|
|
Kaper JB, Nataro JP, Mobley HL (2004). Pathogenic Escherichia coli. Nature Review Microbiology 2(2):123-140.
Crossref
|
|
|
|
|
Kayisoglu S, Yilmaz I, Demirci M, Yetim H (2003). Chemical composition and microbiological quality of the doner kebabs sold in Tekirdag market. Food Control 14:469-474.
Crossref
|
|
|
|
|
Kim JY, Kim SH, Kwon NH, Bae WK, Lim JY, Koo HC, Kim JM, Noh KM, Jung WK, Park KT, Park YH (2005). Isolation and identification of Escherichia coli O157:H7 using different detection methods and molecular determination by multiplex PCR and RAPD. Journal of Veterinary Science 1:7-19.
Crossref
|
|
|
|
|
Kumar HS, Karunasgar I, Karunasagar I, Teizou T, Shima K, Yamasaki S (2004). Characterization of Shiga toxin producing Escherichia coli (STEC) isolated from seafood and beef. FEMS Microbiology 233:173-178.
Crossref
|
|
|
|
|
Lupindu AM (2017). Epidemiology of shiga toxin-producing Escherichia coli 0157:H7 in Africa in review. South African Journal of Infectious Diseases 33(1):24-30.
Crossref
|
|
|
|
|
Mean PS, Slutsker L, Dietz V, McCaig LF, Bresee JS, Shapiro C (2015). Food-related illness and death in the United States. Emerging Infectious Diseases 5:607-625
Crossref
|
|
|
|
|
Momtaz H, Jamshidi A (2013). Shiga toxin-producing Escherichia coli isolated from chicken meat in Iran: Serogroups, virulence factors, and antimicrobial resistance properties. Poultry Science 92:1305-1313.
Crossref
|
|
|
|
|
Najand LM, Khalilli M (2007). Detection of shiga-like toxigenic Escherichia coli from raw milk cheeses produced in Kerman lran. Veterinarski Arhiv 77:515-522.
|
|
|
|
|
Nakazawa M, Akiba M (2001). Swine as a potential reservoir of Shiga toxin-producing Escherichia coli 0157:H7 in Japan. Emerging Infectious Diseases 5:833-844.
Crossref
|
|
|
|
|
Oksuz O, Arici M, Kurultay S, Gumus T (2004). Incidence of Escherichia coli 0157:H7 in raw milk and white pickled cheese manufactured from raw milk in Turkey. Food Control 15:453-456.
Crossref
|
|
|
|
|
Onanuga A, Igbeneghu O, Lamikanra A (2014). A study of prevalence of diarrhoeagenic E. coli in children from Gwagwalada federal Capital Territory, Nigeria. The Pan African Medical Journal 17(3):146-152.
Crossref
|
|
|
|
|
Perera A, Clarke CM, Dykes GA, Fegan N (2015). Characterization of shiga toxigenic Escherichia coli O157 and non-O157 isolates from ruminant feces in Malaysia. Biomed. Research International, pp.1-8.
Crossref
|
|
|
|
|
Siriken B, Pamuk S (2004). Investigation of the incidence of E. coli 0157:H7 and L. monocytogenes from ground beef sold in Afyon district. I. National Veterinary Food Hygiene Congress, Ankara Uni. Vet. Fac., pp. 101-109.
|
|
|
|
|
Siriken B, Pamuk S, Ozakin C, Gedikoglu S, Eyigor M (2004). The investigation of Salmonella spp. and Escherichia coli 0157:H7 from Turkish Soudjouck in Afyon district. National Veterinary Food Hygiene Congress, Veterinarian - Ankara Üniversitesi Veteriner Fakültesi 29:147-155.
|
|
|
|
|
United States Department of Agriculture (USDA) (2007). Multistate Outbreak of E. coli O157 Infections Linked to Topp's Brand Ground Beef Patties.
|
|
|
|
|
Vijayan C, Ajaykumar VJ, Bhattacharya A Bhanurekka V (2017). Detection of enterohaemorrhagic E. coli 0157: H7 from beef and chevon sold in and around Puducherry. Journal of Entomology and Zoology Studies 5(6):1395-1403.
|
|
|
|
|
Vinothkumar A, Pal UK, Mandal PK, Das CD, Antony PX, Thanislass J (2014). Detection of Shigatoxigenic Escherichia coli in beef in Puducherry region by PCR and antimicrobial efficacy of cardamom and star anise powders against it. Journal of Meat Science 3(2):60-65.
|
|
|
|
|
Yusuf Y, Abdulmalik BS, Aliyu MI, Ummukulthum LH, Raymond JN (2018). Risk of Shiga Toxigenic Escherichia coli 0157:H7 infection from raw and fermented milk in Sokoto Metropolis, Nigeria. Journal of Pathogens 9(3):1-5.
Crossref
|
|
|
|
|
Zahraa AJ, Saad SF, Manal AH, Bashar QK (2016). Detection of Escherichia coli 0157:H7 in food. World Journal of Experimental Sciences 2:83-86.
|
|