ABSTRACT
Yeasts are very important in food production by affecting the quality and safety of different food products. Yeasts are commonly used in the production of beer, wine and bread; but many yeast species have been isolated from different African fermented foods. The present study aims to determine the biodiversity of yeasts isolated from selected Nigerian fermented foods. A combination of phenotypic and molecular tests were employed in the identification of yeasts. A total of 145 yeasts were isolated from six different food products. The yeasts belong to six genera namely Saccharomyces, Candida, Cyberlindnera, Meyerozyma, Trichosporon and Galactomyces. The most frequently encounter was Saccharomyces cerevisiae followed by Candida glabrata. The present study has confirmed the biodiversity of yeasts isolated from fermented food of Nigerian origin.
Key words: Phenotypic characterization, molecular, indigenous yeasts, Nigeria.
In different parts of the world, humans consumed many types of fermented food products. Traditional knowledge is used in the processing of available plant and animal materials to produce fermented food (Achi, 2005). Fermentation is the chemical modification of foods by enzymes of living microorganisms. During the production of fermented foods, microorganisms that are present in the raw materials or added as starter culture change the raw materials both biochemically and organoleptically into edible products that are culturally acceptable to the maker and consumer.
In Nigeria different types of nutrient rich crops are processed by fermentation into foods or used as condiments (Iwuoha and Eke, 1996). Most of these food crops are not consumed without fermentation because of the presence of certain toxic components or antinutritional factors. When foods are fermented a general improvement takes place in the shelf life, texture, taste, aroma as well as nutritional value. The fermentation process is usually performed at the household level or at a small scale. The different types of traditional fermented food products in Nigeria include fufu, gari, ogi and nono.
Fufu and gari are fermented cassava product while ogi is produced from different types of cereals such as maize, sorghum and millet (Omemu et al., 2007). Nono is a fermented milk product. The traditional fermented alcoholic beverages include the palm wine, burukutu and agadagidi. Palm wine is produced from the sugary sap of the African oil palm (Elaeis guineensis) and the raphia palm (Raphia hookeri) (Sanni and Lonner, 1993).
Burukutu is an alcoholic beverage produced from red sorghum (Sorghum bicolor) while agadagidi is produced from ripe plantain pulp (Sanni and Lonner, 1993). The production of these food products is by spontaneous fermentation and therefore different types of microbes are usually found. The microorganisms commonly found in Nigerian traditional fermented foods and alcoholic beverages are yeasts and lactic acid bacteria. However, yeasts are responsible for the alcoholic content of the beverages.
The fermentation of carbon sources by yeasts for production of alcoholic beverages is the oldest and most economically important of all biotechnologies. In order to maximise alcoholic yield and maintain sensory quality, it is necessary to select suitable yeast strains for alcoholic fermentation. The occurrences of yeasts in significant number ranging from 105 to 108 cfu/g in many traditional African fermented foods have been reported (Greppi et al., 2013). Most of the previous studies on identification of yeasts from Nigerian traditional fermented food products used cultural, physiological and biochemical tests for yeast strains identification (Sanni and Lonner, 1993; Omemu et al., 2007; Chilika et al., 2010; Umeh and Okafor, 2016 and Olowonibi, 2017). These methods of identification are laborious and most importantly often fails to identify strains correctly to the species level. DNA sequence based methods provide a rapid and accurate means of microbial identification and permit a certain level of phylogenetic classification of the species into genera or even families (El-Sharoud et al., 2009). The main objective of the current study is to identify yeasts isolated from some Nigerian traditional fermented food products.
Sample collection
Ten samples each of traditional fermented food products like retted cassava (fufu) (FF), ogi-baba (OBB), burukutu (BKT), agadagidi (AGG) and palm wine (PAW) were collected from Ibadan Metropolis immediately after production in sterile containers and transported to the laboratory for immediate use in the isolation of yeasts. Agadagidi was prepared in the laboratory by simulating the traditional method.
Isolation of yeasts
The different food samples were used in the preparation of ten-fold serial dilutions using sterile distilled water. Then 0.1ml of the serial dilutions was spread on potato dextrose agar (PDA) that has been supplemented with 100 mg/ml of chloramphenicol to inhibit the growth of bacteria. The yeast cultures obtained were then purified by repeated streak-inoculation on PDA. The pure cultures obtained were stored under refrigeration until needed for further studies (Kurtzman et al., 2011).
Identification of yeast isolates
Microscopic examination of yeast isolates
The pure cultures of the yeasts were streaked onto thin sterile agar and the cells were examined with a Leitz Ortholux phase-contrast microscope (Leica Microsystems, Wetzlar, Germany) and the images were recorded electronically with a Sony XC-75CCD camera.
Phenotypic identification of yeast isolates
Physiological and biochemical tests were performed by fermentation and assimilation tests. Different carbon compounds such as hexoses, pentoses, polysaccharides, alcohols and organic acids were used. The nitrogen compounds include sodium nitrate, sodium nitrite, ethylamine, lysine and cadaverine (Kurtzman et al., 2011).The carbon compounds used in the study include glucose,sucrose,raffinose,melibiose,galactose,lactose,trehalose,maltose,melezitose,salicin,sorbose, rhamnose,xylose, L-arabinose, D-arabinose and ribose.The alcohol used in this test includes methanol, ethanol, 2-propanol, 1-butanol, glycerol, erythritol, ribitol, xylitol, galacitol, mannitol, glucitol and inositol. The yeasts were grouped together based on their phenotypic properties and representatives were selected for DNA sequencing.
Molecular identification.
DNA sequence analysis
Whole cells grown on YM agar (1.0% glucose, 0.5% peptone, 0.3% malt extract, 0.3% yeast extract, 2.0% agar) for 1 to 3 days were suspended to a density of 1+ in water (Lachance et al., 1999). Ten microlitres of the suspension was incorporated into 20 µL amplification reactions. The D1 and D2 domains of the large subunit ribosomal RNA were amplified using primers NL1 (52-GCATATCAATAAGCGGAGGAAAAG) and NL4 (52-GGTCCGTGTTTCAAGACGG) (Kurtzman and Robnett, 1998). Amplification by the polymerase chain reaction was conducted following the instructions provided by the supplier of heat-activated Taq polymerase (in vitrogen), in the presence of 1.5mM MgCl2 in an M.J Research PTS 200 cycler. The mixture was held for 2 min at 95°C and then subjected to 35 cycles at 94°C for 15 s, annealing temperature for 15 s, and 72°C for 40 s, with final extension for 5 min at 72°C. The annealing temperature varied from 55.7 to 53.7°C during the first 20 cycles and kept constant for the remainder. The amplified DNAs were concentrated and cleaned on Qiagen spin columns, and sequenced with ABI sequencer (Applied Biosystems) at the John P Robarts Research Institute, London, Ontario, Canada. The sequences were edited and aligned with the program Mega, version 5.0 and compared with published sequences (Kurtzman and Robnett, 1998).
The yeasts isolated from differen fermented food products are presented in Table 1. Saccharomyces cerevisiae were isolated from all the food products analysed in this study with the exception of nono. S. cerevisiae is the most frequently isolated with highest number of occurrence. Other yeasts were isolated from one, two or three of the food products studied. S. cerevisiae have been isolated from different fermented food products such as palm wine, dolo, kpete-kpete, tchoukoutou and so on (Glover et al., 2009; Kayode et al., 2011; Ngoc et al., 2013; Djegui et al., 2015; Olowonibi, 2017).
The yeasts isolated in this work exhibited different cellular shapes. The majority had ovoid shapes as shown in Plate 1. The other shape exhibited by some is cylindrical as shown in Plate 2. The yeasts have been reported to exhibit different shapes such as oval and cylindrical as reported by Kurtzman et al. (2011).
The yeasts were identified by sequencing as species of seven different genera (Figure 1) namely Saccharomyces, Candida, Cyberlindnera, Pichia, Meyeromyces, Trichosporon and Galactomyces. The species that were isolated are S. cerevisiae. Sanni and Lonner (1993) reported the isolation of eleven strains of S. cerevisiae from some Nigerian traditional alcoholic beverages like palm wine, burukutu, agadagidi and sekete. Also, Sefa-Dedeh et al. (1999) isolated eight strains of S. cerevisiae from traditional brewing of pito in Ghana.
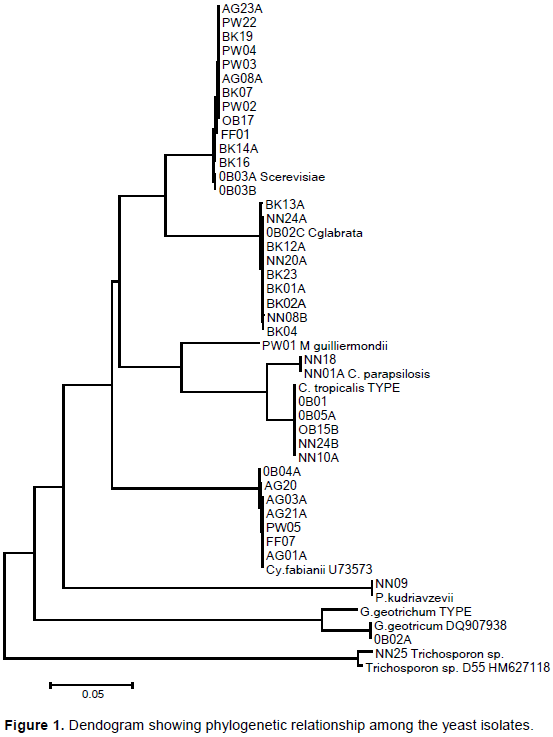
Moreover, Greppi et al. (2013) reported on the diversity of yeasts in ogi, mawe, gowe and tchoukoutou in Benin Republic. S. cerevisiae was isolated from ogi and mawe. Apart from S. cerevisiae another group of yeast commonly encounter in Nigerian traditional fermented food are members of the genus Candida. Candida glabrata effectively a species of Nakaseomyces a relative of the genus Saccharomyces was also isolated. Candida glabrata has been isolated from Zimbabwean opaque beer among other yeasts (Misihairabgwi et al., 2015). Candida tropicalis was another yeasts isolated from the fermented food products. C. tropicalis was characterized from tchapalo, a traditional sorghum beer from Coted’Ivoire.Candida parapsilosis was also isolated from the fermented food products. A study of yeasts in a small scale gari production in Nigeria gave rise to M. guilliermondii, P kudriavzevii (synonym Candida krusei) and two relatives of the genus Pichia, C. inconspicua and C. rugopelliculosa (Oguntoyinbo, 2008).
Pichia kudriavzevii was also isolated from the fermented food products. Occurrence of Pichia kudriavzevii in Ghanaian nunu was reported by Akabanda et al. (2013). P. kudriavzevii has also been isolated from Nigerian palm wine (Nwaiwu et al., 2016). Pichia spp. has also been reportedly isolated from Nigerian palm wine (Chilaka et al., 2010). Satuurnispora (Pichia) saitoi and Wickerhamomyces anomala (Pichia anomala) were isolated during fermentation of cassava for fufu production. Meyerozyma (Pichia) caribbica was isolated from local food crops in Nigeria (Ebabhi, 2013).
Cyberlindnera fabianii was also isolated from some of the fermented foods studied. Cyberlindnera fabianii have been isolated from the microbiota of fermented masau fruits in Zimbabwe (Irma et al., 2017). Trichosporon sp was also isolated from the fermented food studied. Pedersen et al. (2012) reported the occurrence of Trichosporon asahii from fura-a West African fermented cereal product. Galactomyces geotrichum was also isolated from the fermented food products. Galactomyces geotrichum has been listed as among the yeasts that occur in African fermented food products in a review by Johansen et al. (2019). Therefore, this present work has confirmed the biodiversity of yeasts that are associated with Nigerian traditional fermented food products. The next line of our research will be to investigate the technological applications of these yeasts.
The authors have not declared any conflict of interests.
The authors appreciate an award of fellowship by Canadian Commonwealth Scholarship for graduate student exchange program which was used to carry out this work in Canada.
REFERENCES
|
Achi OK, Ukwuru M (2015). Cereal-based fermented foods of Africa as functional foods. International Journal of Food Microbiology and application 2(4):71-83.
|
|
|
|
Akabanda F, Owusu-Kwarteng J, Tano-Debrak K, Glover RLK, Nielsen DS, Jespersen L (2013). Taxonomic and molecular characterization of lactic acid bacteria and yeasts in nunu, a Ghanaian fermented milk product. Food Microbiology 34(2):277-283.
Crossref
|
|
|
|
|
Chilaka CA, Uchechukwu N, Obidiegwu JE, Akpor OB (2010). Evaluation of the efficiency of yeasts isolates from palm wine in diverse fruit wine production. African Journal of Food Science 4(12): 764-774.
|
|
|
|
|
Djegui KY, Kayode APP, Tokpohozin ES, Gachomo EW, Kotchoni SO, Hounhouigan JD (2015). Phenotypic characters of yeasts isolated from kpete-kpete, a traditional starter of a Benin opaque sorghum beer. African Journal of Biotechnology 14(27):2227-2233.
Crossref
|
|
|
|
|
Ebabhi AM, Adekunle AA, Okunnowo WO, Osuntoki AA (2013). Isolation and characterization of yeast strains from local food crops. Journal of Yeast and Fungal Research 4(4):38-43.
|
|
|
|
|
El-Sharoud WM, Belloch C, Peris D, Querol A (2009). Molecular identification of yeasts associated with traditional Egyptian dairy products. Journal of Food Science 74 (7):M341-M346.
Crossref
|
|
|
|
|
Glover RLK, Sawadogo-Lingani H, Diawara B, Jespersen L, Jakobsen M (2009). Utilization of Lactobacillus fermentum and Saccharomyces cerevisiae as starter cultures in the production of "dolo". Journal of Applied Biosciences 22:1213-1319.
|
|
|
|
|
Greppi A, Rantsiou K, Padonou W, Hounhouigan J, Jespersen L, Jakobsen M, Cocolin L (2013). Determination of yeast diversity in ogi, mawe, gowe and tchoukoutou by using culture-dependent and -independent methods. International Journal of Food Microbiology 165(2):84-88.
Crossref
|
|
|
|
|
Iwuoha CI, Eke OS (1996). Nigerian indigenous fermented foods: their traditional process operation, inherent problems, improvements and current status. Food Research International 29(5-6):527-540.
Crossref
|
|
|
|
|
Johansen PG, Owusu-kwarteng J, Parkouda C, Padonou SW, Jespersen L (2019). Occurrence and impotant of yeasts in indigenous fermented food and beverages produced in Sub-Sahara Africa. Frontiers in Microbiology 10:1-22.
Crossref
|
|
|
|
|
Kayode APP, Vieira-Dalode G, Linnemann AR, Kotchoni SO, Hounhouigan AJD, van Boekel MAJS, Nout MJR (2011). Diversity of yeasts involved in the fermentation of tchoukoutou, an opaque sorghum beer from Benin. African Journal of Microbiology Research 5(18):2737-2742.
Crossref
|
|
|
|
|
Kurtzman CP, Robnett CJ (1998). Identification and phylogeny of ascomycetous yeasts from analysis of nuclear large subunit (26S) ribosomal DNA partial sequences. Antonie Van Leeuwenhoek 73(4):331-371.
Crossref
|
|
|
|
|
Kurtzman CP, Fell JW, Boekhout T, Robert V (2011). Methods for isolation, phenotypic characterization and maintenance of yeasts. Elsevier pp 87-110.
Crossref
|
|
|
|
|
Lachance MA, Bowles JM, Starmer WT, Barker JSF (1999). Kodamaea kakaduensis and Candida tolerans, two new yeast species from Australian Hibiscus flowers. Canadian Journal Microbiology 45(2):172-177.
Crossref
|
|
|
|
|
Misihairabgwi JM, Kock L, Pretorious E, Pohl C, Zvauya R (2015). Characterization of yeasts isolated from traditional opaque beer beverages brewed in Zimbabwean households. African Journal of Microbiology Research 9 (8):549-556.
Crossref
|
|
|
|
|
Ngoc NM, Minh NP, Dao DTA (2013). Isolation and characterization of yeast strains for palm (Borasscus flabelliffer) wine fermentation. International Journal of Engineering Research and Technology 2(11):3602-3615.
|
|
|
|
|
Nwaiwu O, Ibekwe VI, Amadi ES, Udebuani AC, Nwanebu FC, Oguoma OI, Nnokwe JC (2016). Evaluation of fermentation products of palm wine yeasts and role of Saloglottis gabonensis supplement on products abundance. Beverages 2(9):13-20.
Crossref
|
|
|
|
|
Oguntoyinbo FA (2008). Evaluation of diversity of Candida species isolated from fermented cassava during traditional small scale gari production in Nigeria. Food Control 19(4):465-469.
Crossref
|
|
|
|
|
Olowonibi OO (2017). Isolation and characterization of palm wine strains of Saccharomyces cerevisiae potentially useful as bakery yeasts. European Journal of Experimental Biology 7(11):1-13.
Crossref
|
|
|
|
|
Omemu AM, Oyewole OB, Bankole MO (2007). Significant of yeasts in the fermentation of maize for ogi production. Food Microbiology 24(6):571-576.
Crossref
|
|
|
|
|
Pedersen LL, Owusu-Kwarteng J, Thorsen L, Jespersen L (2012). Biodiversity and probiotic potential of yeasts isolated from fura, a West Africa spontaneously fermented cereal. International Journal of Food Microbiology 159(2):144-151.
Crossref
|
|
|
|
|
Sanni AI, Lonner C (1993). Identification of yeasts isolated from Nigerian traditional alcoholic beverages. Food Microbiology 10:517-523.
Crossref
|
|
|
|
|
Sefa-Dedeh S, Sanni AI, Tetteh G, Sakyi-Dawson E (1999). Yeasts in the traditional brewing of pito in Ghana. World Journal of Microbiology and Biotechnology 15(5):593-597.
Crossref
|
|
|
|
|
Umeh SO, Okafor JNC (2016). Isolation, characterization and identification of yeast (Saccharomyces cerevisiae) from three local beverage drinks. International Journal Series in Multidisciplinary Research 3(5):44-55.
|
|
|
|
|
Van Rijswijck IM, Irma MHR, Martijn FLD, Tjakko A, Dick K, Eddy JS (2017). Genome sequences of Cyberlindnera fabianii 65, Pichia kudriavzevii 129 and Saccharomyces cerevisiae 131 isolated from fermented masau fruits in Zimbabwe. Genome Announcement 5(14):e00064-17.
Crossref
|
|