ABSTRACT
Emissions of biological contaminants (microbes) from intensive pig farming may cause environmental problems due to lack of proper waste management. This work was conducted to assess bacteriological pollution of soil in pig farm and to detect the presence of antibiotic resistance gene of the prevailing bacteria. Soil samples were collected from March to August 2013. The method included bacterial enumeration (10 -1 to 10 -8) in Nutrient, Xylose Lysine Deoxycholate (XLD), Eosin Methylene Blue (EMB) and MacConkey agars. Bacteria were identified using API 20E test kit; antibiotic susceptibility test were also determined and identification of resistance gene was carried out using molecular procedures. The viable cells in soil samples ranged from 0 to 2.44 × 1010 cfu/ml. Pseudomonas luteola, Salmonella choleraesuis spp arizonae, Escherichia coli 1, Enterobacter aerogenes, etc. were the predominant isolates. Sixty-seven percent of isolates were resistant to Penicillin G while 79% were resistant to Spectinomycin. The resistance genes detected in most isolates were Van A, InuA, Sul2, blaTEM and Otr B. The results showed that bacterial pathogens isolated from pig farm soil were not only diverse but also possessed multiple Antibiotic Resistance Gene (ARG) and this may have possible dire consequences on the environment and public health.
Key words: Microbes, pathogens, pollution, antibiotics, resistance gene, environment.
Mishandling of pig farm waste and animal droppings may impact negatively on the physical environment, especially polluting the soil with bacteriological pathogens. The pollution may consequentially cause serious waterborne and airborne diseases by either as a result of ingestion or direct contact, or inhalation of contaminated aerosols (Ramírez et al., 2005). Applying animal waste to the soil may solve waste disposal issues; unfortunately this can also introduce bacterial pollutants to the soil, groundwater systems, and surface water in the surrounding environment (Obasi et al., 2008). Potential sources of bacterial pollution in pig farms include feedlot pastures,
treatment lagoons, manure storage, and also land application fields (Hong et al., 2013).
Pathogens can be transported in soil receiving waste through movement with infiltrating water, and surface run-off water and with the movement of sediments and waste particles (Jamieson et al., 2002). The bacteria that are already at the soil surface can act as suspended particles thus trapping even more bacteria which are deposited into the soil (Jamieson et al., 2002). Therefore, the bacteria that are deposited into the soil can travel with mobile water and their cells can interact with air or solid phase, which result in temporary or permanent immobilization (Åuczkiewicz and Quant, 2007). In such conditions, bacteria can also be entrapped in stagnant pore water between gas bubbles (Åuczkiewicz and Quant, 2007). During rain events, connectivity between mobile and immobile water increases, allowing bacteria to migrate with the advancing wetting front, thus allowing the bacteria to reach the ground water through absorption and infiltration and thus contaminate the groundwater (Åuczkiewicz and Quant, 2007).
Agricultural run-offs or seepage of pig farm waste are known to be critical sources of bacterial pollution in soil (Tyrrel and Quinton, 2003). Due to high use of antibiotics in pig farm as growth promoters and also to treat infections and diseases some of these antimicrobial agents are excreted with pig’s faeces in an unaltered state (Hong et al., 2013). This is because antibiotics are poorly absorbed into the gut of farm animals and therefore can be exposed to natural faunas and floras of soil through faeces and urine of pigs and in soil through surface runoff of pig farm seepage (Kumar et al., 2005). Some of the bacterial pathogens are able to resist the antibiotics in the gut of pigs and can be exposed to soil during defecation, thus introducing bacteria that possesses antibiotic resistance gene (ARG) in the environment (Obasi et al., 2008).
The rapid growth of antimicrobial agents in the environment as a result of the extensive use in pig farms emphasizes the need for intervention (Roberts, 2005). Most antibiotic residues cannot be removed by wastewater treatment plants and these residual antibiotics can be released into the environment and may exert selection pressure on natural soil microorganisms (Heuer et al., 2011). Furthermore, soil may receive inputs of antimicrobials, which can serve to amplify antibiotic resistance genes (ARGs) (Chee-Sanford et al., 2009; Heuer et al., 2011). The use of antibiotics agents can also cause overgrowth of bacterial expressing a resistance gene to the antibiotic agent and therefore may aggregate the evolution of complex genetic vectors encoding, expressing, linking, and spreading the bacteria and other resistance genes (Cheng et al., 2013). In order to further establish the contribution of agricultural practices to the environment, this study was conducted to assess the bacteriological dynamics of soil environment from a pig farm and to detect the presence of antibiotic resistance gene of the prevailing bacteria.
Study area
The research was conducted at the pig feedlot section situated about 25 km south of Pretoria (25°52′S 28°13′E / 25.867°S 28.217°E / -25.867; 28.217) in Gauteng, South Africa. The section of the farm where this work was conducted housed about 105 pigs, that is, about 70 sows, 10 boars and 25 piglets. The pig farm was divided into sections A and B. The pigs in Section A were kept in semi-intensive range where their enclosure floor was not cemented and therefore were in direct contact with the soil. Section B pigs were housed in intensive unit, namely: Farrowing house, grower house, dry sows house and weaning house. Soil samples were taken from section A of the pig feedlot.
Soil sampling
Top soil samples were collected at the Agricultural Research Council - Animal Production Institute (ARC-API) pig farm section. These samples were collected monthly from March to August 2013 between 07. 00 and 09. 00 hours am on weekly basis. Soil samples of about 2 kg were collected in sterile polythene bags using soil auger at depth of 30 cm (Bhat et al., 2011). The soil samples were collected from 5 sites in the pig farm, that is, pig farm enclosures (Enc S), soil samples from 20 m (Enc S-20 m) and 100 m (Enc S-100 m) away from the pig farm enclosures, soil samples from 20 m (CW S-20 m) and 100 m (CW S-100 m) away from pig farm constructed wetland used for the treatment of pig farm wastewater. Soil samples were placed on ice in a cooler box immediately after sampling and transported to the lab to be analyzed.
Bacteria isolation
Bacteria isolation procedure was adapted from the methods of Ramirez et al. (2005). One hundred milliliter of sterile distilled water was poured into a sterile conical flask (200 mL) and 10 g of the soil sample was weighed and added to the distilled water in the bottle. The flask was tightly capped and mixed thoroughly for 30 min using magnetic stirrer plate. Serial dilution method was adapted where 1 mL of the soil sample supernatant was used in performing the serial dilution from 10-1 up to 10-8 using sterile 0.9 % (w/v) saline solution (Bezuidenhout et al., 2002). About 1 mL of each dilution was added to 15 mL of agar in test tube, mixed thoroughly and the contents were poured into a petri-dish, allowed to solidify and incubated at 37°C for 2 days. The media that were used included Nutrient agar, MacConkey Agar (for isolation of Shigella spp., Salmonella spp., Yersinia spp., Providencia spp., Serratia spp.). Xylose Lysine Deoxycholate agar (XLD agar) was used for isolation of Enterobacter spp., Proteus spp., Pseudomonas spp., and Eosin Methylene Blue (EMB) was used for isolation of Escherichia coli, Aerobacter aerogenus, Citrobacter spp., and Klebsiella spp.. Viable cells were counted after 2 days of incubation from each petri-dish and isolates were picked and streaked three times on nutrient agar for pure colony. Serial dilutions were done in triplicates
Identification of isolates
Isolates were identified using Analytical Profile Index (API 20E) identification kit (bioMérieux South Africa (Pty) Ltd). Pure isolates were streaked on nutrient agar and incubated at 37°C for 24 h. The overnight grown cultures were then inoculated in 5 mL of 0.85% (w/v) saline solution and the turbidity of the resulting solution was adjusted to 0.5 McFarland Standard. The manufacturer procedure was followed in inoculating the isolates on the Analytical Profile Index (API 20E) test strips. All reactions were read according to the recommendations of the manufacturer and the seven-digit octal number was calculated, and the organism identity was determined using the apiweb software.
Antimicrobial assays
Antibiotic susceptibility/resistance was determined by the Kirby-Bauer disk diffusion method according to standard procedure outlined by Clinical and Laboratory Standards Institute (CLSI) (CLSI, 2007) and Kumar et al. (2013). The isolates were screened for susceptibility/resistance to a panel of antibiotics using Mueller Hinton agar (Oxoid, UK). Antibiotics tested were Penicillin G (10 µg), Sulphamethaxazole (25 µg), Vancomycin (30 µg), Ampicillin (10 µg), Amoxicillin (25 µg), Apramycin (15 µg), Neomycin (30 µg), Tilmocosin (15 µg), Oxytetracyclin (30 µg), Spectinomycin (25 µg), Lincomycin (15 µg), Trimethoprim (2.5 µg), Nalidixic Acid (30 µg), Gentamycin (10 µg), Tetracycline (30 µg), Ceftadizime (10 µg), Norflaxacin (10 µg), and Nitrofurantoin (300 µg). These antibiotics are used at the pig farm to treat diseases and for growth promotion. Most antibiotics for growth promotions are added to feeds and are given on a regular basis and pigs at the age of 7 months are given a dosage range of 10 to 40 g of growth promoter antibiotics.
E. coli ATCC 25922, Ps. aeruginosa ATCC 19429, and S. marscensce ATCC 14041 were used as controls. All Antibiotic susceptibility/resistance tests for isolates were assayed in triplicates and incubated at 37°C for 20 h. The Multidrug Resistance Index (MDRI) of each sample was estimated using the following equation: , where A represents the aggregate antibiotic resistance score of all isolates from the sample, B represents the number of antibiotics, and C represents the number of isolates from the sample (Krumperman, 1983).
Detection of resistance gene in identified isolates
DNA isolation
The isolates were cultured in nutrient broth and incubated for 24 h at 37°C. NucleoSpin Tissue Genomic DNA purification kit (Machery-Nagel, Germany) was used to isolate genomic DNA from the identified isolates. The manufactures procedure was followed for isolation of the genomic DNA (support protocol for bacteria). The purity and yield of the DNA was assessed spectrophotometrically (NanoDrop ND-2000c, Thermo) by calculating the A260/A280 ratios and the A260 values to determine protein impurities and DNA concentrations.
Polymerase chain reaction (PCR) amplification for detection resistance genes
PCR amplification assays were performed to detect the presence of antimicrobial resistance genes in identified bacterial isolates. The method adopted was that outlined by Hsu et al. (2007). The sequences of 29 primers used for PCR amplification of antibiotic resistance genes are listed in Table 1. Amplification of the DNA was performed in a PCR apparatus with iProof High Fidelity DNA Polymerase (BIO-RAD). The 20 µL reaction mixture contained 0.02U/ µL iProof DNA Polymerase; 1X iProof HF Buffer; 3% DMSO; 700 µM MgCl2; 200 µM dNTPs; 0.5 µM Forward Primer; 0.5 µM Reverse Primer; 1 µg DNA Template and 11.4 µL of nuclease free water. The PCR cycling conditions were as follows: 98°C for 30 s, followed by 35 cycles at 98°C for 10 s, 30 s at the annealing temperature of Primer, 72°C for 30 s, and termination at 72°C for 10 min. Pseudomonas. aeruginosa ATCC 19429 were used as control and a reaction mixture with no DNA template was used as blank. Amplified DNA from each sample (10 µL) was mixed with 1 µL of 6x loading buffer dye and loaded on a 1 % horizontal agarose gel containing 0.5 mg/ mL of ethidium bromide. A 100 bp DNA ladder ranging from 100 to 3000 bp (Thermo Scientific) was also added on each gel to confirm the size of amplified DNA bands. All gels were run in 1 X TAE buffer at 5 V cm-1 for 30 min, and visualized by UV trans-illumination.

Statistical analysis
Calculation of means and standard deviations for viable counts were performed using Microsoft Excel office 2010 version. Test of significance (two-way ANOVA) were performed using SPSS 17.0 version for Windows program (SPSS, Inc.). All tests of significance were considered statistically significant at P values of < 0.05.
Bacteria population
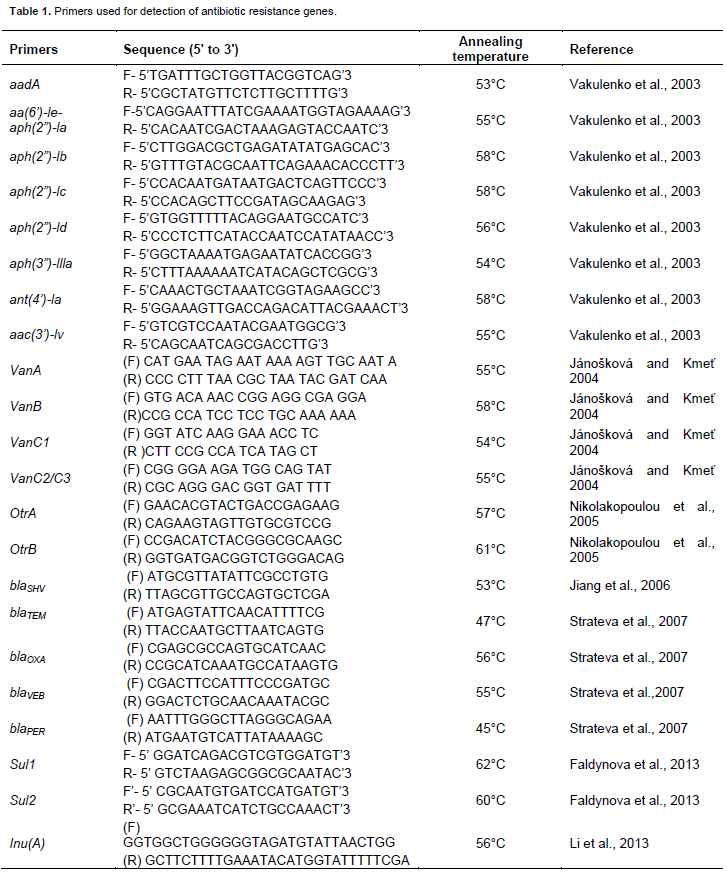
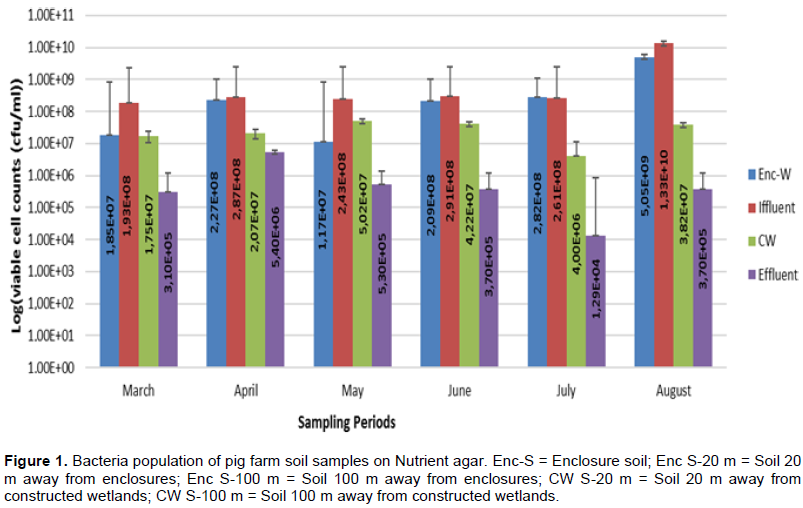
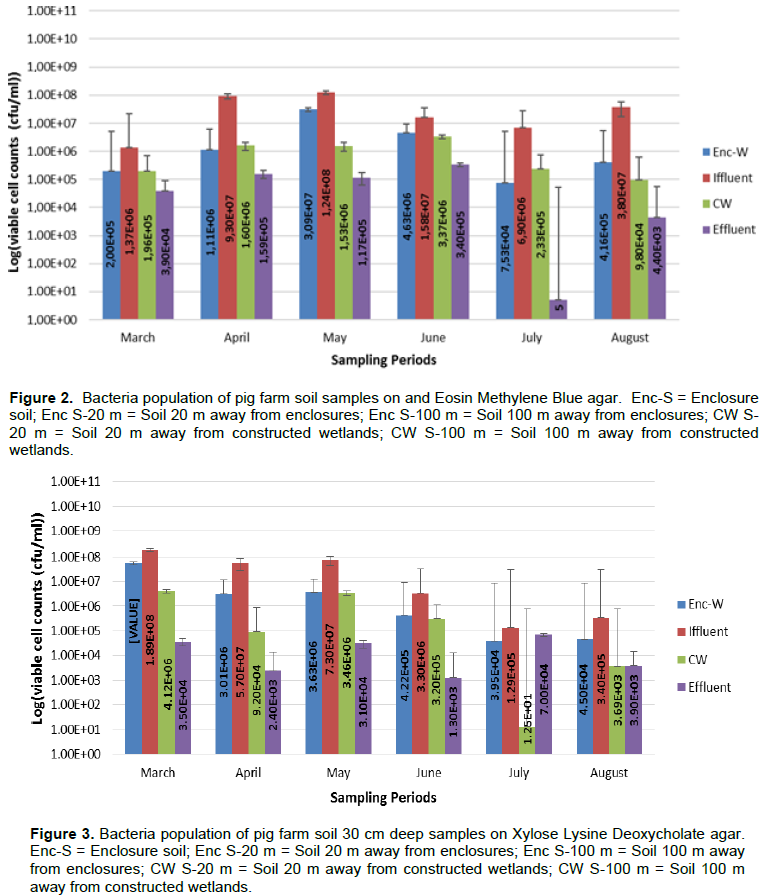
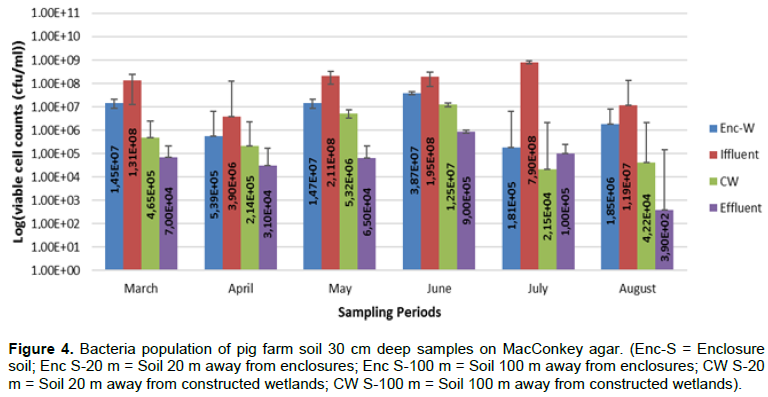
Results for viable cell counts of pig farm soil samples are shown in Figures 1 to 4. In Nutrient agar (Figure 1), the viable cells ranged from 1.29 × 104 to 1.33 × 1010 cfu/mL, and the results did not reveal any significant variation between months and also between sampling points. On EMB agar (Figure 2), the viable cell counts ranged from 5.00 × 100 to 1.24 × 108 cfu/mL. The results for viable cell on EMB agar did not show significant variation between months and also between sampling points. The viable cell counts ranged from 1.25 × 101 to 1.89 × 108 cfu/mL in XLD agar (Figure 3). The results for viable cell counts on XLD agar varied significantly (p<0.05) monthly and did not show significant difference between sampling points. The viable cells ranged from 3.90 × 102 to 7.90 × 108 cfu/mL in MacConkey agar (Figure 4). The results varied significantly (p<0.05) monthly and insignificantly at sampling points.
Bacteria identification
The identification of 49 isolates using API20E kit gave the following species: Pseudomonas Luteola, Salmonella choleraesuis spp arizonae, Escherichia coli 1, Enterobacter aerogenes, Pasteurella pneumotropica, Proteus vulgaris, Pseudomonas Aeruginosa, Burkholderia cepacia, Stenotrophomonas maltophilia, Shwenella putrefaciens, Klebsiela pneumonia, Serratia liquefaciens, Enterobacter sakaziki, Citrobacter braakii, Enterobacter amnigenus 2, Enterobacter amnigenus 1, and Serratia marcescens.
The analysis for susceptibility, using 18 different antibiotics, revealed resistant (R), susceptible (S) and intermediate (I) isolates to tested antibiotics are shown in Figure 5. The results showed that 67% of isolates were resistant to penicillin G, 70 % to vancomycin, 70% to oxytetracycline and 79% to spectinomycin. Among the isolates, 88% were susceptible to norflaxacin 95% to ceftadizime, 72% to Tetracycline, and 60% to nitrofurantoin. In addition, 58% of the isolates were susceptible to neomycin and 51% to nalidixic acid. With ampicillin and gentamycin, the percentage of susceptible isolates (44 and 44% respectively) when compared to those that were resistant to both ampicillin and gentamycin (51 and 49% respectively) did not vary greatly.

The detection of resistance gene showed that most isolates had VanA, VanB, OtrA, OtrB resistance genes (Table 3). The resistant genes; ant (4’)-la. AadA, ant (4’)-la, and blaVEB; were not detected in all isolates and only Burkholderia cepacia and Enterobacter amnigenus 1 had aph (2”)-lc. Only E. coli 1 had aph (2”)-ld resistance gene. Only Salmonella choleraesuis spp arizonae had VanD resistance gene. As well, only Serratia liquefaciens had VanC resistance gene and only Enterobacter amnigenus 1 had InuD resistance gene. The gel electrophoresis results for detection of InuA resistance gen is shown in Figure 6.

The results for phenotypic antibiotic resistance and Multidrug Resistance Index are shown in Table 2. The most predominate phenotype multiple resistance were P-RL-VA-AML-APR-AMP-TIL-OT-SH-MY-NI and P-RL-VA-AML-APR-AMP-N-TIL-OT-SH-MY with 5.1%, respectively, as phenotype antibiotics resistance percentage. About 45 isolates had more than 5 phenotype antibiotic resistance patterns where Penicillin G (P), Sulphamethaxazole (RL), Vancomycin (VA), Ampicillin (AML), Apramycin (AMP), Amoxicillin (APR), Oxytetracyclin (OT), Spectinomycin (SH) were the most predominant.
Bacterial contamination has negative impacts on the environment as bacterial pathogens can compete with indigenous soil microorganism for nutrients, transfer antimicrobial resistance genes to indigenous soil microorganism important for soil remediation (Sasákováet al., 2007). Results in this study for viable cell counts were lower as compared to those observed by Cook et al. (2010) where from 2.58 × 1010 to 1.49 × 1011 cfu/mL were obtained from farrowing facilities at pig farm.
Salmonella choleraesuis spp. arizonae and E. coli 1 were detected in the soil samples from pig enclosures, soil 20 m away from enclosures and soil 20 m away from constructed wetland treating pig farm wastewater. Although S. choleraesuis spp arizonae presence was confirmed in all soil samples examined, this was occasional as it was not detected in the months of June and July. Still even their small count should be alarming because they can easily spread under favorable conditions and make a serious source of environmental pollution which can lead to airborne diseases (Cook et al., 2010). In addition, the identities of the isolates especially E. coli 1 was not performed to strain level and, therefore, the isolate could belong to the serotype E. coli 0157:H4 and E. coli 0104:H4 that have been reported to cause disease in humans (Muniesa et al., 2012). Detection of these bacterial pathogens in soil samples in pig farm may be attributable to the high load of animal excreta in the pig farm seepage and serves as an indicator for possible bacteriological pollution and may have an effect on the soil ecological balance and aquatic life (Ezeronye and Ubalua, 2005). The results obtained in this study was also similar to that observed by Tymczyna et al. (2000) where bacteria such as Salmonella spp, Klebsiella spp, Pseudomonas spp, Proteus spp, Enterobacter aerogenes, Citrobacter spp. etc. were isolated from soil samples from pig farm enclosures and surrounding environment.
The phenotype resistance patterns observed in this study showed that the isolates were highly resistant to more than three antibiotics and the multidrug resistance index (MDRI) of isolates was also high (Table 2). Among 49 phenotype multiple resistance patterns observed, the phenotypes that were mostly observed were P-RL-VA-AML-APR-AMP-TIL-OT-SH-MY-NI, and P-RL-VA-AML-APR-AMP-N-TIL-OT-SH-MY. Some of resistance patterns were not frequently detected, and some of the isolates were found to be resistant to only 1 antimicrobial agent. Multidrug Resistance Index was observed in this study where 5 isolates had an MDRI ranging from 66 to 83% and 8 isolates had MDRI of 61%. In a study conducted by Kotzamanidis et al. (2009), it was observed an AML-CAZ-VA-TE as the most occurring phenotype pattern in isolates from pig farm environment, whereas in this study the most occurring phenotype patterns were P-RL-VA- AML-AMP-SH-MY and P-RL-VA-AML-APR-SH-MY. The phenotype pattern, also, differed from those observed by Kainer et al. (2007) where the most occurring phenotype pattern was P-RL-VA-APR-N-TIL-OT-SH-MY. However, Werner et al. (2008) observed phenotype patterns VA-SH-TM and SH-MY-TM which were also detected in this study.
The result observed in this study for antibiotic susceptibility/resistance test (Figure 4) showed that these organisms have been well exposed to the tested antibiotics and they have developed mechanisms to evade or avoid the effects of these antibiotics. This high antimicrobial resistance is of concern because antibiotic resistance genes can be transferred from pathogens to non-pathogenic (indigenous) microorganism. These observations were also similar to those observed by Kainer et al. (2007) and Werner et al. (2008) where resistance to similar antibiotics was reported. The increased use of antibiotics in livestock industry can introduce a selective pressure which leads to the development of resistance or even multi-resistance characteristics in some of the bacterial
populations (Chen and Jiang, 2014).
Since soil bacteria like Burkholderia Cepacia are usually used in bioremediation of soil, the acquiring of antibiotic resistance gene renders these soil bacteria unsafe for bioaugmentation application (Krista et al., 1996). Bacterial pathogens, such as E. coli, Proteus spp., Salmonella spp., Enterobacter spp., were observed to have multiple resistance genes to most of the antibiotics tests. Contamination of the natural environment by these bacteria may accelerate the growth of algae, deplete dissolved oxygen in water systems, cause eutrophication, and emit toxins that can kill fish and other animals (Pandey et al., 2014).
The results showed that most isolates possessed aa (6’)-le-aph (2”)-la gene, aph (3”)-llla genes for aminoglycosides resistance, Sul1 gene and Sul2 gene for Sulphamethaxazole resistance, VanA, VanB and VanC2/C3 resistance genes for vancomycin, Inu A and Inu C resistance genes for lincomycin, OtrA and OtrB resistance genes for oxytetracyclines and blaTEM and blaPE resistance gene for beta-lactamase resistance. AadA, ant (4’)-la, and blaVEB resistant genes were not detected in all isolates and only Burkholderia Cepacia and Enterobacter amnigenus 1 had aph (2”)-lc. Only E. coli 1 had aph (2”)-ld resistance gene, only Salmonella choleraesuis spp arizonae had VanD resistance gene, only Serratia liquefaciens had VanC resistance gene and only Enterobacter amnigenus 1 had InuD resistance gene. Enterobacter amnigenus 1 and Burkholderia Cepacia were the only 2 isolates to possess aph (2”)-lc resistance gene, while Enterobacter sakaziki and E. coli 1 were the only 2 isolates to poses blaSHV resistance gene.
Sul resistance genes have been reported as the most frequently detected ARGs in pig farm seepage by Zhu et al. (2013). Equally, from this study, Sul resistance gene (Table 3) was detected in most of isolates from soil samples from pig farm. In a study conducted by Chee-Sanford et al. (2001), OtrA resistance gene was the most abundant and detected in surface soil (depth between 0-30 cm) in pig farm and this was consistent with the current observations where OtrA resistance gene was observed in most surface soil sample isolates.
In a study conducted by Faldynova et al. (2013), sul1 and aadA resistance genes were very abundant in pig farm soil and seepage while sul2 resistance gene was less observed in isolates from pig farm surrounding environment. The observations from this study differed from those observed by Faldynova et al. (2013) because aadA resistance gene was not detected in all isolates and sul1 and sul2 resistance genes were more or less equally observed in isolates from soil samples in pig farm. In a study conducted by Li et al. (2013), they observed an abundance of InuF resistance gene in all soil samples collected in pig farm and this was not the case in this study as InuF resistance gene was less detected in soil samples from pig farm. Similar results reported by Li et al. (2013) were also reported by Cheng et al. (2013) where widespread of InuF resistance gene was also observed in soil samples proximal to pig farm.
This study revealed that the soil at pig farm was contaminated with bacterial pathogens as bacteria such as E. coli, E. aerogenes, K. pneumonia, P. vulgaris etc. were detected especially with their ARGs in all soil samples from pig farm. The presence of this bacterial pathogens in soil sampled in the pig farm in this study was observed not only to be diverse but were also abundant and this may threaten the quality of the surrounding natural environment. In addition, this study also revealed that this prevailing bacterial pathogens isolated from pig farm soil had multiple ARG. The presence of ARG in soil causes horizontal gene transfer and may form a critical zone for the species-rich environmental microbiota and antibiotic-resistant microorganism to exchange genetic material with indigenous soil microorganism (Schulz et al., 2012).
The authors have not declared any conflict of interests.
REFERENCES
|
Bezuidenhout CC, Mthembu N, Puckree T, Lin J (2002). Microbiological evaluation of the Mhlathuze River, Kwazulu-Natal (RSA). J. Water SA. 28(3):281-286.
Crossref
|
|
|
|
Bhat SH, Darzi AB, Dar MS, Ganaie MM, Bakhshi SH (2011). Correlation of soil physico-chemical factors with vam fungi distribution under different agroecological conditions. Int. J. Pharm. Biol. Sci. 2(2):98-107.
|
|
|
|
|
Chee-Sanford JC, Aminov RI, Krapac IJ, Garrigues- Jeanjean N, Mackie RI (2001). Occurrence and diversity of tetracycline genes in lagoons and groundwater underlying to swine production facilities. Appl. Environ. Microbiol. 67(4):1494-1502.
Crossref
|
|
|
|
|
Chee-Sanford JC, Mackie RI, Koike S, Krapac IG, Lin YF, Yannarell AC (2009). Fate and transport of antibiotic residues and antibiotic resistance genes following land application of manure waste. J. Environ. Qual. 38(3):1086-1108.
Crossref
|
|
|
|
|
Chen Z, Jiang X (2014). Microbiological safety of chicken litter or chicken litter-based organic fertilizers: a review. Agriculture 4(1):1-29.
Crossref
|
|
|
|
|
Cheng W, Chen H, Su C, Yan S (2013). Abundance and persistence of antibiotic resistance genes in livestock farms: A comprehensive investigation in eastern China. J. Environ. Int. 61(3):1-7.
Crossref
|
|
|
|
|
Clinical and Laboratory Standards Institute (CLSI) (2007). Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M7-A6. Wayne, PA, USA: Clinical and Laboratory Standards Institute.
|
|
|
|
|
Cook K, Rothrock Jr. MJ, Lonanh N, Sorrell JK, Loughring JH (2010). Spatial and temporal changes in the microbial community in an anaerobic swine waste treatment. Anaerobe 16(2):74-82.
Crossref
|
|
|
|
|
Ezeronye OU, Ubalua AO (2005). Studies in the effect of abattoir and industrial effluents on the heavy metals and microbial quality of Aba River in Nigeria. Afr. J. Biotechnol. 4(3):266-272.
|
|
|
|
|
Faldynova M, Videnska P, Havlickova H, Sisak F, Juricova H, Babak V, Steinhauser L, Rychlik I (2013). Prevalence of antibiotics resistance genes in faecal samples from cattle, pigs, and poultry. J. Vet Med. 58(6):298-304.
|
|
|
|
|
Heuer H, Schmitt H, Smalla K (2011). Antibiotic resistance gene spread due to manure application on agricultural fields. J. Curr. Opin. Microbiol. 14(3):236-243.
Crossref
|
|
|
|
|
Hong P, Yannarell AC, Dai Q, Ekizoglu M, Mackie RI (2013). Monitoring the pertubation of soil and groundwater microbial communities due to pig production activities. J. Appl. Environ. Microbiol. 79(8):2620-2629.
Crossref
|
|
|
|
|
Hsu WB, Wang JH, Chen PC, Lu YS, and Chen JH (2007). Detecting low concentrations of Shigella sonnei in environmental water samples by PCR. Microbiol. Lett. 270(2):291-298.
Crossref
|
|
|
|
|
Jamieson RC, Gordon RJ, Sharples KE, Stratton GW, Madani A (2002). Movement and persistence of fecal bacteria in agricultural soils and subsurface drainage water: A review. Can. Biosys. Eng. 44:1-19.
|
|
|
|
|
Jánošková A, KmeÅ¥ V (2004). Vancomycin resistance genes in Enterococcus spp. Strains isolated from alpine accentor and chamois. J. Microbiol. 73(2):211-214.
Crossref
|
|
|
|
|
Jiang X, Zhang Z, Li M, Zhou D, Ruan F, Lu Y (2006). Detection of extended-spectrum beta-lactamases in clinical isolates of Pseudomonas aeruginosa. J. Antimicrob Agents Chemother. 50(7):2990-2995.
Crossref
|
|
|
|
|
Kainer MA, Devasia RA, Jones TF, Simmons BP, Melton K, Chow S, Broyles J, Moore KL, Craig AS, Schaffner W (2007). Response to emerging infection leading to outbreak of linezolid-resistant enterococci. J. Emerg. Infect. Dis. 13(7):1024-1030.
Crossref
|
|
|
|
|
Kotzamanidis C, Zdragas A, Kourelis A, Moraitou E, Papa A, Yiantzi V, Pantelidou C, Yiangou M (2009). Characterization of VanA type Enterococcus faecium isolates from urban and hospital wastewater and pigs. J. Appl. Microbiol. 107(3):997-1005.
Crossref
|
|
|
|
|
Krista DL, Kerr A, Jones MC, Caracciolo JA, Eskridge B, Jordan M, Miller S, Hughes D, King N, Gilligan PH (1996). Accuracy of four commercial systems for identification of Burkholderia cepacia and other gram-negative nonfermenting bacilli recovered from patients with cystic fibrosis. J. Clin. Microbiol. 34(4):886-891.
|
|
|
|
|
Krumperman PH (1983). Multiple antibiotics resistance indexing of Escherichia coli to identify high-risk sources of fecal contamination of foods. J. Appl. Environmen. Microbiol. 46(1):165-170.
|
|
|
|
|
Kumar K, Gupta SC, Chander Y, Singh AK (2005). Antibiotic use in agriculture and its impact on the terrestrial environment. J. Adv. Agron. 87(2):1-54.
Crossref
|
|
|
|
|
Kumar S, Tripathi VR, Garg SK (2013). Antibiotic resistance and genetic diversity in water-borne Enterobacteriaceae isolates from recreational and drinking water sources. Int. J. Environ. Sci. Technol. 10(4):789-798.
Crossref
|
|
|
|
|
Li L, Sun J, Liu B, Zhao D, Ma J, Deng H, Li X, Hu F, Liao X, Liu Y (2013). Quantification of lincomycin resistance genes associated with lincomycin residues in waters and soils adjacent to representative swine farms in China. J. Environ. Contamin. Tox. 4:364-372.
Crossref
|
|
|
|
|
Åuczkiewicz A, Quant B (2007). Soil and groundwater fecal contamination as a result of sewage sludge land application. Pol. J. Environ. Stud. 16(4):587-593.
|
|
|
|
|
Muniesa M, Hammerl JA, Hertwig S, Appel B, Brüssow H (2012). Shiga Toxin-Producing Escherichia coli O104:H4: A New Challenge for Microbiology. J. Appl. Environ. Microbiol. 78(12):4065-4073.
Crossref
|
|
|
|
|
Nikolakopoulou TL, Egan S, van Overbeek LS, Guillaume G, Heuer H, Wellington EMH, van Elsas JD, Collard J, Smalla K, Karagouni AD (2005). PCR detection of oxytetracycline resistance genes otrA and otrB in tetracycline-resistant Streptomycete isolates from Diverse Habitats. J. Curr. Microbiol. 51(4):211-216.
Crossref
|
|
|
|
|
Obasi LN, Nwadinigwe CA, Asegbeke JN (2008). Study of trace heavy metal in fluted pumpkin leaves grown on soil treated with sewage sludge and effluents. Proceedings 31st International Conference of C.S.N Petroleum Training Institute (PTI) Conference Centre Complex Warri, Pp. 241-244.
|
|
|
|
|
Pandey PK, Kass PH, Soupir ML, Biswas S, Singh VP (2014). Contamination of water resources by pathogenic bacteria. AMB Express 4:51-67.
Crossref
|
|
|
|
|
Ramírez G, Martínez R, Herradora M, Castrejón F, Galvan E (2005). Isolation of Salmonella spp. from liquid and solid excreta prior to and following ensilage in ten swine farms located in central Mexico. J. Bioresour. Technol. 96(5):587-595.
Crossref
|
|
|
|
|
Roberts MC (2005). Update on acquired tetracycline resistance genes. FEMS Microbiol. Let. 245(2):195-203.
Crossref
|
|
|
|
|
Sasáková N, Juriš P, Papajová I, Vargová M, Venglovský J, OndrašoviÄová O, OndrašoviÄ M (2007). Bacteriological and parasitological risks associated with agricultural wastewaters and sewage subjected to biological treatment. XIII International Congress in Animal Hygiene, Pp. 985-989.
|
|
|
|
|
Schulz J, Friese A, Klees S, Tenhagen BA, Fetsch A, Rösler U, Hartung J (2012). Longitudinal Study of the Contamination of Air and of Soil Surfaces in the Vicinity of Pig Barns by Livestock-Associated Methicillin-Resistant Staphylococcus aureus. J. Appl. Environ. Microbiol. 78(16):5666-5671.
Crossref
|
|
|
|
|
Strateva T, Ouzounova-Raykova V, Markova B, Todorova A, Marteva-Proevska Y, Mitov I (2007). Widespread detection of VEB-1-type extended spectrum β-lactamases among nosocomial ceftazidime-resistant Pseudomonas aeruginosa isolates in Sofia, Bulgaria. J. Chemother. 19(2):140-145.
Crossref
|
|
|
|
|
Tymczyna L, Chmielowiec-Korzeniowska A, Saba L (2000). Bacteriological and parasitological pollution of the natural environment in the vicinity of pig farm. Pol. J. Environ. Stud. 9(3):209-214.
|
|
|
|
|
Tyrrel SF, Quinton JN (2003). Overland flow transport of pathogens from agricultural land receiving faecal wastes. J. Appl. Microbiol. 94:87-93.
Crossref
|
|
|
|
|
Vakulenko SB, Donabedian SM, Voskresenskiy AM, Zervos, MJ, Lerner ST, Chow JW (2003). Multiplex PCR for the detection of aminoglycoside resistance genes in enterococci. J. Antimicrob. Agents Chemother. 47(4):1423-1426.
Crossref
|
|
|
|
|
Werner G, Coque TM, Hammerum AM, Hope R, Hryniewicz W, Johnson A (2008). Emergence and spread of vancomycin resistance among enterococci in Europe. J. Eur. Surv. 13(47):256-367.
|
|
|
|
|
Zhu YG, Johnson TA, Su JQ, Qiao M, Guo GX, Stedtfeld RD (2013). Diverse and abundant antibiotic resistance genes in Chinese swine farms. Proc. Natl. Acad. Sci. 110(9):3435-3440.
Crossref
|
|