Full Length Research Paper
ABSTRACT
Fresh fruits are popular sources of healthy diets with low energy density. Since they are consumed raw, it may act as a source of foodborne disease and a reservoir for antibiotic-resistant organisms. This study aimed to determine microbial prevalence among the fruits sold around hospital along with antimicrobial profiles. Thirty-five different types of fruits were bought from around Dhaka Medical College and Hospital (DMCH) and analyzed for the presence of bacteria. Antibiotic sensitivity, detection of ESBL, AmpC β-lactamase, and MBL positive strains were done by standard methods followed by PCR to detect ESBL, AmpC β-lactamase and MBL genes. Twenty-seven different organisms were isolated: Klebsiella spp. (33.33%), Citrobacter spp. (29.64%), Enterobacter spp. (22.22%), Escherichia coli (11.11%) and Staphylococcus aureus (3.70%). Among them, 48.15% were resistant to different antibiotics. Only one organism (Citrobacter spp.) produced ESBL phenotypically (7.69%). Two (15.38%) were positive for AmpC β-lactamase and one of these (Enterobacter spp.) possessed SHV and CTX-M15A genes by PCR. Imipenem resistance was 84.62% of the antibiotic-resistant organisms and 10 (90.91%) were phenotypically MBL positive. By PCR, one Enterobacter spp. had MBL encoding gene OXA-48. Fresh fruits, contaminated with pathogens, might be a source of resistant organisms' transmission and contribute to public health issues.
Key words: Antibiogram, bacteria, Bangladesh, fresh fruits, fruit venders around hospital.
INTRODUCTION
Fresh fruits are good sources of vitamins, minerals, phytonutrients and dietary fiber. It is also considered as a measure to decrease heart diseases and some cancers (Xyli et al., 2019). Moreover, fruits are regarded microbiologically safer than other unprocessed foods. Thus, to promote fruits and fresh products intake, the Food and Agriculture Organization (FAO) of the United Nations and the World Health Organization (WHO) recommended a minimum consumption of 400 g of vegetables and fruits per day (Hölzel et al., 2018). But, fruit surfaces are not free from microorganisms, and generally consumed raw which can impose foodborne diseases (Rinc?n and Neelam, 2021). As a result, fresh fruits can act as a means of human exposure to antibiotic-resistant bacteria containing antibiotic resistance genes (Liu and Song, 2019). These antibiotic resistant bacteria containing antibiotic resistance genes on the fruit or vegetable surfaces give rise to risks regarding environmental health ecologically (Sun et al., 2021).
Since β-lactams, specially extended spectrum cephalosporins and carbapenems are the drugs of choice to treat resistant gram negative bacteria, β-lactamases, such as penicillinase, extended-spectrum β-lactamases (ESBLs), cephalosporinases (AmpC) and carbapenemases have emerged worldwide (Ye et al., 2018). This leads to increase use of the last line antibiotic, the pivotal alternative, colistin, which ultimately gives rise to colistin-resistant bacteria, in particular, through horizontal transfer of mcr gene (Bakthavatchalam et al., 2018).
Biswas et al. (2020) demonstrates diverse gram negative bacteria on the surfaces of fresh fruits and vegetables, among which Vibrio spp., Lactobacillus spp., Pseudomonas spp. and Salmonella spp. are dominant. The transfer of these organisms to fresh fruits and vegetables may occur at multi-stage from production to home kitchen- during production through the use of animal manure and contaminated irrigation water, during the post-harvest stage, transport, conservation and processing by handlers (Richter et al., 2019). It certainly poses a potential public health threat since they are able to exchange resistance genes with intestinal bacteria during their colonization and passage through the intestines leading to further dissemination in the environment (van Hoek et al., 2015).
Previously, chromosomally encoded MBL enzymes or chromosomally mutated genes responsible for colistin resistance was not transferable, hence were clinically negligible (Anyanwu et al., 2020; Elshamy and Aboshanab, 2020). Afterwards, various plasmid-encoded carbapenemase genes, such as Imipenem-resistant Pseudomonas-type carbapenemases (IMP), Verona integron-encoded MBL (VIM), New Delhi MBL (NDM) and mobilized colistin gene (mcr) have been described which are capable of horizontal transfer (Liu et al., 2016; Elshamy and Aboshanab, 2020). Recently, ESBL, cephalosporinase, carbapenemase and mcr gene-producing gram-negative bacteria isolated from fresh vegetables and fruits have been reported in several studies (Ye et al., 2018; Liu and Song, 2019; Yang et al., 2019; Manageiro et al., 2020) which is very alarming.
Therefore, in this study, we determined the extent of bacterial contamination and an anti- microbial profile of the organisms isolated from the fruit surfaces, sold and consumed in and around DMCH.
MATERIALS AND METHODS
This cross sectional study was conducted at the department of Microbiology, Dhaka Medical College, Dhaka from April 2021 to September 2021 in 5 phases at different time. Thirty five different raw fruits, namely, five apples, five mangoes, five guava, five sets of blackberry, five bunches of grapes, five oranges, four pineapples, and one sugarcane juice, were bought from the fruit venders around Dhaka Medical College and Hospital, Dhaka. Samples were transported to the laboratory carefully in sterile polythene bags for bacteriological analysis (Leff and Fierer, 2013). Fruits were washed with 20 ml sterile distilled water and fluid was transferred into separate beaker. A ten-fold serial dilution of the samples was performed in the Trypticase Soya broth (TSB).
Determination of total viable count (TVC) by bacterial enumeration
Bacterial enumeration was used to determine the number of colony forming units (CFUs) (Angela et al., 2010).
Isolation and identification of bacteria
After incubating the diluted samples in TSB for 24 h, they were subcultured in blood agar and MacConkey agar media followed by incubation at 37°C for 48 h. A single colony was further subcultured until pure culture was obtained. Identification of bacteria was performed on the basis of colony morphology, Gram’s staining reaction and biochemical tests (catalase, coagulase, sugar utilization, gas and H2S production in Triple Sugar Iron agar media, citrate utilization test in Simmon’s Citrate agar, motility, indole and urease production in Motility Indole Urea agar media) (Cheesbrough, 2009).
Antimicrobial susceptibility test
Susceptibility to antimicrobial agents of all isolated organisms was determined by modified Kirby-Bauer technique using Mueller-Hinton agar media. Zones of inhibition were interpreted according to CLSI guidelines (CLSI, 2021). FDA guideline was followed for tigecycline (“Antimicrobial Susceptibility test Interpretive Criteria”, 2022).
Detection of ESBL producers by double disc synergy test
30µg ceftazidime disc and amoxiclav (20 µg + 10 µg) disc were placed 20 to 25 mm apart (center to center) in the Mueller- Hinton agar plate and was observed for clear extension of the edge of zone of inhibition of cephalosporin disc towards amoxyclav disc after incubating at 37°C for 24 h (Iqbal et al., 2017).
Phenotypic detection of carbapenemase producers
Organisms were considered carbapenemase producers if positive for any of the following methods.
Double disc synergy (DDS) test
After inoculating the test inoculums (compared with McFarland standard) in Mueller- Hinton agar plates, imipenem disc was placed on the inoculated plate along with a blank disc containing 20 µl of Tris- EDTA and 20 µl of 1:320 diluted 2- mercaptopropionic acid, placed 10 mm apart. A clear extension of the edge of the inhibition zone of imipenem disc towards Tris- EDTA- MPA disc was observed after incubating at 37°C for 24 h (Kim et al., 2007).
Combined disc (CD) assay
Two imipenem discs (one supplemented with 5 µl of 0.5 M EDTA solution containing approximately 930 µg EDTA) were placed on an inoculated Mueller- Hinton agar plate following incubation at 37°C for 24 h. An increased zone of diameter of ≥6 mm around the disc containing imipenem supplemented with EDTA compared to the disc containing imipenem only was interpreted as MBL producer (Qu et al., 2009).
Modified Hodge test (MHT)
A lawn culture of 1:10 dilutions of 0.5 McFarland’s standard Escherichia coli ATCC 25922 broth was done on a Mueller- Hinton agar plate. A 10 µg imipenem disc was placed in the center of the plate. Then 0.5 McFarland’s standard were made by three imipenem resistant gram negative organisms and streaked from the edge of the disc to the periphery of the plate in three different directions. After overnight incubation, the presence of clover leaf shaped zone of inhibition was interpreted as MHT positive (Amjad et al., 2011).
Detection of anti-microbial resistance genes by PCR
DNA extraction
300 μl distilled water was mixed with bacterial pellets and vortexed until mixed well. Then DNA was extracted in block heater (DAIHA Scientific, Seoul, Korea) at 100°C for 10 min for boiling followed by cooling on ice pack and centrifugation at 4°C at 13500 g for 10 min. The extracted DNAs were kept at -20°C for future use (Farzana et al., 2022).
Amplification through thermal cycler
PCR was performed in a DNA thermal cycler (Eppendorf AG, Mastercycler gradient, Hamburg, Germany) after mixing of mastermix and primers with DNA template. Each PCR run comprised of preheat at 94°C for 10 min followed by 36 cycles of denaturation at 94°C for 1 min, annealing at specified temperatures for 45 s, extension at 72°C for 1 min with final extension at 72°C for 10 min. Gel electrophoresis was done in 1.5% agarose (Bethesda Research Laboratories). DNA bands were detected by staining with ethidium bromide (0.5 μl/ ml) for 30 min at room temperature and visualized with UV transilluninator (Gel Doc, Major Science, Taiwan).
Data analysis
The result of the study was recorded systematically. Data analysis was done by using ‘Microsoft Office Excel 2010’ program and according to the objectives of the study. The test of significant was calculated by using X2 test. P value < 0.05 was taken as minimal level of significance.
RESULTS
Fruits were collected on five occasions and 35 cultures were done. Among those 35 fruits 25 (71.43%) yielded growth of 27 different gram positive and gram negative organisms, two fruits showed growth of double organisms. Ranges of microbial count in apple was 1×102 to 6×102 CFU/ml, guava was 3×102 to 3×103 CFU/ml, blackberry was 0.1×10 to 3×102 CFU/ml, orange was 0.3×102 to 2.4×102 CFU/ml, mango was 1.2×102 to 8×102 CFU/ml, grape was 1×102 to 1.4×103 CFU/ml and pineapple was 1.6×102 CFU/ml, and sugar cane juice was 2×102 CFU/ml.
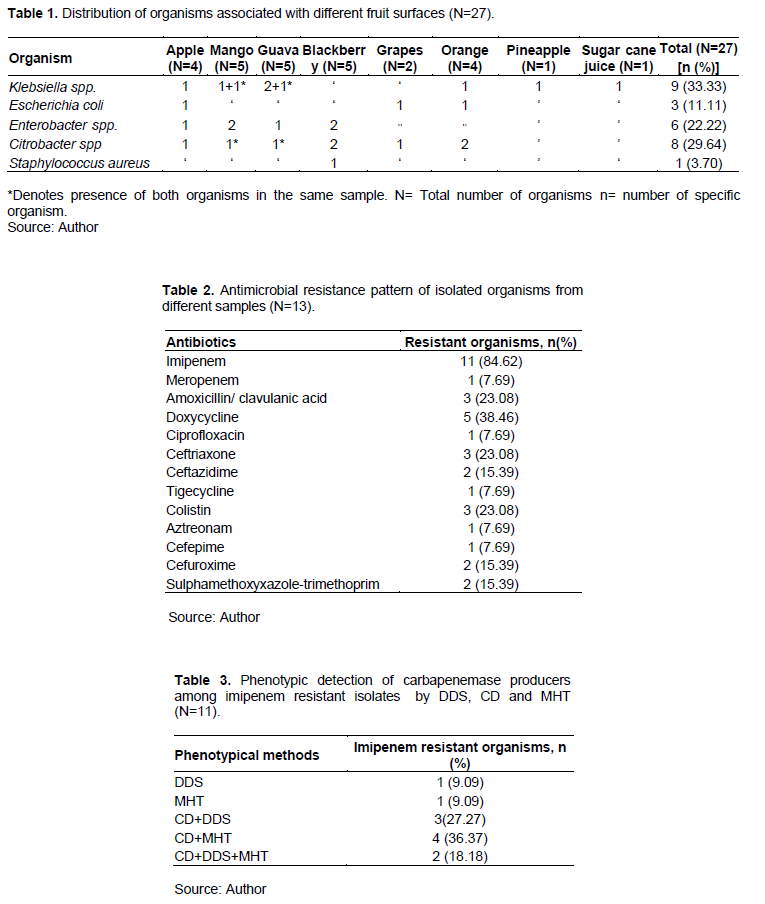
Among 27 different organisms, nine (33.33%) were Klebsiella spp., eight (29.64%) were Citrobacter spp., six (22.22%) were Enterobacter spp., E. coli were three (11.11%) and lastly Staphylococcus aureus were one (3.70%) (Table 1).
Fourteen (51.85%) organisms were sensitive to all drugs, and 13 (48.15%) organisms were found to have resistance to different antibiotics.
Four out of six Enterobacter spp. and two of the nine Klebsiella spp. were multidrug resistant. Within the resistant organisms, only one (7.69%) was extended spectrum-β-lactemase (ESBL) producer (Citrobacter spp.) and two (15.38%) were AmpC-β-lactemase producers (Klebsiella spp. and Enterobacter spp.) phenotypically. One of the AmpC-β-lactemase producers (Enterobacter spp.) was positive for both SHV and CTX-M15A gene (Table 2).
Eleven (84.62%) out of 13 antibiotic resistant organisms were imipenem resistant and all of them were phenotypically detected as carbapenemase producers by CD test, DDS test or MHT. Ten (90?91%) of them were phenotypically positive for MBLs (Table 3), considering one that was only positive for MHT. Among the ten MBL positive samples, only one (Enterobacter spp.) was positive for OXA-48 gene. No MBL positive organisms were positive for VIM, NDM-1, NDM-2, IMP, OXA-181, OXA-10, KPC genes. The only organism which was resistant to tigecycline, was not positive for tetA gene.
DISCUSSION
Fruits are consumed in raw state which facilitates the transfer of microorganisms on the fruit surfaces to human along with antimicrobial resistance genes (Chelaghma et al., 2021). Bacterial enumeration identified in this study highlighted the fact that fresh fruits contaminated with pathogenic organisms can act as a transmission vehicle for human diseases (Biswas et al., 2020). It can cause a lot of sufferings for the patients, like a prolonged hospital stay, which ultimately inflects the treatment cost of the patients.
The microbial load of the fruits used in this study varied with the types of food, but their presence and anti-microbial patterns highlighted the fact that, fruits could be contaminated with pathogenic bacteria and thus may act as a vehicle for transmitting diseases.
In a previous study, among 105 fruit samples, 126 bacterial isolates were identified (different species of Enterobacteriaceae and Staphylococcus spp.) (Al-Kharousi et al., 2016). In another study, among 25 fruit samples, 106 bacterial isolates were identified (Escherichia coli, Salmonella spp., Vibrio spp., Bacillus spp. and Staphylococcus spp.) (Sarker et al., 2018). In this study, 27 isolates of six different species among 35 samples were identified. These contaminations may occur from the soil through manure fertilization, or by human through direct contamination (Hölzel et al., 2018). Some studies also confirmed the effect of wastewater for irrigation playing a pivotal role in contaminating fresh farm products such as fresh vegetables and fruits (Adegoke et al., 2018). The difference between the findings may be due to the different sample size, geographic area, seasonal variation or different practices in production and transportation of fruits.
In recent years, the use of antibiotics has increased globally and antibiotic resistance genes have been described in all environment including natural and clinical habitats (Bahram et al., 2018; Chng et al., 2020). The antibiotic resistance genes find their way into the soil by animal or poultry manure and sewage sludge, that is organic manure, and then from the soil to the fruit surfaces by soil-plant system via aerosol from the environment (Zhu et al., 2017; Zhang et al., 2019; Wu et al., 2022).
They spread from fruits and vegetables to human via the food chain (Chen et al., 2019). In this study, almost half of the isolates were resistant to one or more antimicrobial groups. Among them 84.62% were resistant to imipenem, followed by 38.46% to doxycycline. 23.08 and 15.39% were resistant to third generation cephalosporin (ceftriaxone and ceftazidime). At present colistin is considered as the antibiotic of last resort and it is used when a bacteria shows resistance to most of the available antibiotics including carbapenems. In the present study, 23.08% gram negative bacilli were resistant to colistin which is a concern. In a study conducted in Bangladesh Agricultural University, Mymensingh, bacterial isolates of guava such as, Escherichia coli, Vibrio spp., and Staphylococcus spp. were found resistant to ampicillin and cephalexin and Salmonella spp. were also resistant to chloramphenicol, ampicillin and cephalexin (Sarker et al., 2018). In 2018, a study on fresh vegetables revealed 83.3% prevalence of ESBL-producing strains (Iseppi et al., 2018). Whereas, in the present study, only one of the resistant isolates (Citrobacter spp) was positive for ESBL phenotypically. Furthermore, in this study, one AmpC β-lactamase producer harbored both SHV and
Furthermore, they are serious threat to public health, as these genes have the potential to be transmitted by horizontal gene transfer (Groussin et al., 2021). Gram negative bacteria isolated from clinical samples from patients of Dhaka Medical College Hospital of Bangladesh showed increased proportion of ESBL producers up to about 2015, after that increasing proportion of MBL producers was observed.
The reason behind it might be due to the fact that ESBL producers are treated by carbapenems and the bacteria have started producing more carbapenemase enzymes due to increased used of carbapenems. Moreover, imported fresh food products seem to be a possible reservoir of carbapenemase- producing gram negative organisms (Solaiman et al., 2021). In a study conducted in Algeria, it reported OXA-48 producing Klebsiella pneumoniae (Touati et al., 2017). In this study, one Enterobacter spp. was found harboring OXA-48.
Wide spread use of antibiotics might contribute in the emergence of multidrug resistant bacteria. Moreover, application of animal manure to the agricultural field using untreated wastewater for irrigation,unhygienic handling at post-harvest stage and during transportation and processing by handlers- all contribute to the spread of drug-resistant bacteria to fresh food products (Richter et al., 2019).
CONCLUSION
A global risk of transmission of multidrug resistant organisms via fresh fruits needs global attention to minimize future public health issues. The findings of this study clearly showed that there was a wide array of micro-organisms in different fruit samples collected from around a tertiary level hospital in Bangladesh; Klebsiella spp. was the most prevalent one. Almost half of the organisms showed resistance to multiple antibiotics and antibiotic resistance genes were found in Enterobacter spp. It is a matter of great concern for the consumers. To reduce the risk for the consumers, improvement of agricultural practice, which includes better water quality for irrigation, safe use of fertilizer, hygienic transportation, should be ensured for the fresh food products including fruits. Moreover, ethical use of antibiotics should be emphasized in all sectors to control antibiotic resistance.
CONFLICT OF INTERESTS
The authors declared no conflicts of interests.
ACKNOWLEDGEMENT
The authors acknowledge the Office of the Director General of Health Services (DGHS), Bangladesh for financial support. They sincerely thank Principal Professor Dr. Md. Titu Miah and vice Principal Dr. Md. Shafiqul Alam Chowdhury of Dhaka Medical College, Dhaka, Bangladesh for their active cooperation.
REFERENCES
|
Adegoke AA, Amoah ID, Stenström TA, Verbyla ME, Mihelcic JR (2018). Epidemiological evidence and health risks associated with agricultural reuse of partially treated and untreated wastewater: a review. Front Public Health 6:337. |
|
|
Al-Kharousi ZS, Guizani N, Al-Sadi AM, Al-Bulushi IM, Shaharoona B (2016). Hiding in Fresh Fruits and Vegetables: Opportunistic Pathogens May Cross Geographical Barriers. In. Merino S (eds.), International Journal of Microbiology. Hindawi Publishing Corporation 2016:4292417. |
|
|
Amjad A, Mirza IA, Abbasi SA, Farwa U, Malik N, Zia F (2011). Modified Hodge test: A simple and effective test for detection of carbapenemase production. Iranian Journal of Microbiology 3(4):189. |
|
|
Angela OE, Ibukunoluwa AO, Oranusi US (2010). Microbial quality of fruits and vegetables sold in Sango Ota, Nigeria. African Journal of Food Science 4(5):291-296. |
|
|
Antimicrobial Susceptibility test Interpretive Criteria: Tigecycline- injection product (2022). U.S Food and Drug Administration. Retrieved from: |
|
|
Anyanwu MU, Jaja IF, Nwobi OC (2020). Occurrence and characteristics of mobile colistin resistance (mcr) gene-containing isolates from the environment: a review. International Journal of Environmental Research and Public Health 17(3):1028. |
|
|
Bahram M, Hildebrand F, Forslund SK, Anderson JL, Soudzilovskaia NA, Bodegom PM, Bengtsson-Palme J, Anslan S, Coelho LP, Harend H, Huerta-Cepas J, Medema MH, Maltz MR, Mundra S, Olsson PA, Pent M, Põlme S, Sunagawa S, Ryberg M, Tedersoo L, Bork P (2018). Structure and function of the global topsoil microbiome. Nature 560(7717):233-237. |
|
|
Bakthavatchalam YD, Pragasam AK, Biswas I, Veeraraghavan B (2018). Polymyxin susceptibility testing, interpretative breakpoints and resistance mechanisms: An update. Journal of Global Antimicrobial Resistance 12:124-136. |
|
|
Biswas B, Azad MAK, Absar N, Islam S, Amin S (2020). Isolation and identification of pathogenic bacteria from fresh fruits and vegetables in Chittagong, Bangladesh. Journal of Microbiology Research 10(2):55-58. |
|
|
Cheesbrough M (2009). District laboratory practice in tropical countries. Part 2. 2nd edn. Cambridge University Press. UK. |
|
|
Chelaghma W, Loucif L, Bendahou M, Rolain JM (2021). Vegetables and fruit as a reservoir of β-lactam and colistin-resistant Gram-negative bacteria: A Review. Microorganisms 9(12):2534. |
|
|
Chen QL, Cui HL, Su JQ, Penuelas J, Zhu YG (2019). Antibiotic resistomes in plant microbiomes. Trends in Plant Science 24(6):530-541. |
|
|
Chng KR, Li C, Bertrand D, Ng AHQ, Kwah JS, Low HM, Tong C, Natrajan M, Zhang MH, Xu L, Ko KKK, Ho EXP, Av-Shalom TV, Teo JWP, Khor CC, Consortium MSUB, Chen SL, Mason CE, Ng OT, Marimuthu K, Ang B, Nagarajan N (2020). Cartography of opportunistic pathogens and antibiotic resistance genes in a tertiary hospital environment. Nature Medicine 26(6):941-951. |
|
|
Clinical and Laboratory Standards Institute (CLSI) (2021). Performance standards for antimicrobial disc susceptibility testing. 31st edition. USA. |
|
|
Elshamy AA, Aboshanab KM (2020). A review on bacterial resistance to carbapenems: epidemiology, detection and treatment options. Future Science OA 6(3):FSO438. |
|
|
Farzana A, Shamsuzzaman SM, Akhter S, Begam M, Jahan H (2022). Detection of Metallo β-lactamases in clinical isolates of Pseudomonas aeruginosa in a tertiary care hospital. Sir Salimulla Medical College Journal 30(1):35-39. |
|
|
Groussin M, Poyet M, Sistiaga A, Kearney SM, Moniz K, Noel M, Hooker J, Gibbons SM, Segurel L, Froment A, Mohamed RS, Fezeu A, Juimo VA, Lafosse S, Tabe FE, Girard C, Iqaluk D, Nguyen LTT, Shapiro BJ, Lehtimäki J, Alm EJ (2021). Elevated rates of horizontal gene transfer in the industrialized human microbiome. Cell 184(8):2053-2067. |
|
|
Hölzel CS, Tetens JL, Schwaiger K (2018). Unraveling the role of vegetables in spreading antimicrobial-resistant bacteria: a need for quantitative risk assessment. Foodborne Pathogens and Disease 15:671-688. |
|
|
Iqbal R, Ikram N, Shoaib M, Muhammad JA, Raja TM, Abid AN, Aanam A, Bushra I, Faiza, N (2017). Phenotypic cofirmatory disc diffusion test (PCDDT), double disc synergy test (DDST), E-test OS diagnostic tool for detection of extended-spectrum β-lactamase (ESBL) producing Uropathogens. Journal of Applied Biotechnology and Bioengineering 3:344-349. |
|
|
Iseppi R, de Niederhäusern S, Bondi M, Messi P, Sabia C (2018). Extended-spectrum β-lactamase, AmpC, and MBL-producing gram-negative bacteria on fresh vegetables and ready- to-eat salads sold in local markets. Microbial Drug Resistance 24:1156-1164. |
|
|
Kim SY, Hong SG, Moland ES, Thomson KS (2007). Convenient test using a combination of chelating agents for detection of metallo-β-lactamases in the clinical laboratory. Journal of Clinical Microbiology 45:2798-2801. |
|
|
Leff JW, Fierer N (2013). Bacterial communities associated with the surfaces of fresh fruits and vegetables. PloS one 8(3):e59310. |
|
|
Liu BT, Song FJ (2019). Emergence of two Escherichia coli strains co-harboring mcr-1 and blaNDM in fresh vegetables from China. Infection and Drug Resistance 12:2627. |
|
|
Liu YY, Wang Y, Walsh PTR, Yi LX, Zhang R, Spencer J, Doi Y, Tian G, Dong B, Huang X, Yu LF, Gu D, Ren H, Chen X, Lv L, He D, Zhou H, Liang PZ, Liu PJH, ShenPJH (2016). Emergence of plasmid-mediated colistin resistance mechanism mcr-1 in animals and human beings in China: a microbiological and molecular biological study. The Lancet Infectious Diseases 16(2):161-168. |
|
|
Manageiro V, Jones-Dias, D, Ferreira E, Caniça M (2020). Plasmid-mediated colistin resistance (mcr-1) in Escherichia coli from non-imported fresh vegetables for human consumption in Portugal. Microorganisms 8(3):429. |
|
|
Mesbah Zekar F, Granier SA, Touati A, Millemann Y (2020). Occurrence of third-generation cephalosporins-resistant Klebsiella pneumoniae in fresh fruits and vegetables purchased at markets in Algeria. Microbial Drug Resistance 26(4):353-359. |
|
|
Qu T, Zhang J, Wang J, Tao J, Yu Y, Chen Y, Zhou J, Li L (2009). Evaluation of phenotypic tests for detection of Metallo-β-lactamase producing Pseudomonas aeruginosa strains in China. Journal of Clinical Microbiology 47(4):1136-1142. |
|
|
Richter L, Du Plessis EM, Duvenage S, Korsten L (2019). Occurrence, identification, and antimicrobial resistance profiles of extended-spectrum and AmpC β-lactamase producing Enterobacteriaceae from fresh vegetables retailed in Gauteng Province, South Africa. Foodborne Pathogens and Disease 16:21-27. |
|
|
Rincón VMV, Neelam DK (2021). An overview on endophytic bacterial diversity habitat in vegetables and fruits. Folia Microbiologica 66(5):715-725. |
|
|
Safain KS, Bhuyan GS, Tasnim S, Hasib SH, Sultana R, Islam MS, Al Mahmud-Un-Nabi M, Sarker SK, Noor FA, Rahat A, Bhuiyan MAM, Islam MT, Manzoor F, Anwar S, Leung D, Qadri SS, Qadri F, Mannoor K (2020). Situation of antibiotic resistance in Bangladesh and its association with resistance genes for horizontal transfer. bioRxiv |
|
|
Sarker MAR, Haque MM, Rifa RA, Ema FA, Islam MA, Khatun MM (2018). Isolation and identification of bacteria from fresh guava (Psidium guajava) sold at local markets in Mymensingh and their antibiogram profile. Vet World 11:1145. |
|
|
Sun Y, Guo G, Tian F, Chen H, Liu W, Li M, Wang S (2021). Antibiotic resistance genes and bacterial community on the surfaces of five cultivars of fresh tomatoes. Ecotoxicology 30(8):1550-1558. |
|
|
Touati A, Mairi A, Baloul Y, Lalaoui R, Bakour S, Thighilt L, Gharout A, Rolain JM (2017). First detection of Klebsiella pneumoniae producing OXA-48 in fresh vegetables from Béja a city, Algeria. Journal of Global Antimicrobial Resistance 9:17-18. |
|
|
van Hoek AHAM, Veenman C, van Overbeek WM, Lynch G, de Roda H, Ana M, Blaak H (2015). Prevalence and characterization of ESBL-and AmpC-producing Enterobacteriaceae on retail vegetables. International Journal of Food Microbiology 204:1-8. |
|
|
Wu J, Wang J, Li Z, Guo S, Li K, Xu P, Ok YS, Jones DL, Zou J (2022). Antibiotics and antibiotic resistance genes in agricultural soils: A systematic analysis. Critical Reviews in Environmental Science and Technology pp. 1-18. |
|
|
Xyli P, Botsaris G, Chrysargyris A, Skandamis P, Tzortzakis N (2019). Variation of microbial load and biochemical activity of ready-to-eat salads in Cyprus as affected by vegetable type, season, and producer. Food Microbiology 83:200-210. |
|
|
Yang F, Shen C, Zheng X, Liu Y, Mohamed MAEGES, Zhao Z, Liao K, Shi Y, Guo X, Zhong R, Xu Z, Tian GB (2019). Plasmid-mediated colistin resistance gene mcr-1 in Escherichia coli and Klebsiella pneumoniae isolated from market retail fruits in Guangzhou, China. Infection and drug resistance 12:385-389. |
|
|
Ye Q, Wu Q, Zhang S, Zhang J, Yang G, Wang J, Xue L, Chen M (2018). Characterization of extended-spectrum β-lactamase producing Enterobacteriaceae from retail food in China. Frontiers in Microbiology 9:1709. |
|
|
Zhang Y-J, Hu H-W, Chen Q-L, Singh BK, Yan H, Chen D, He J-Z (2019). Transfer of antibiotic resistance from manure-amended soils to vegetable microbiomes. Environment International 130:104912. |
|
|
Zhu B, Chen Q, Chen S, Zhu Y-G (2017). Does organically produced lettuce harbor higher abundance of antibiotic resistance genes than conventionally produced? Environment international 98:152-159. |
|
Copyright © 2024 Author(s) retain the copyright of this article.
This article is published under the terms of the Creative Commons Attribution License 4.0