ABSTRACT
The objective of this study was to know the sanitary quality of fish coming from Layo farm. Twenty fishes (Oreochromis niloticus), were selected in four ponds and Escherichia coli were isolated in gills and viscera according to microbiological methods. One hundred and twenty strains of E. coli were isolated, and their virulence was performed by polymerase chain reaction (PCR) using specific primers. Eleven (11) strains (9.16%) including 7 strains of gills (11.66%) and 4 strains of viscera (6.66%) had virulence genes eae, Stx1, Stx2 or ial. Atypical Enteropathogenic E. coli (EPEC, eae+, lack of bfp) was isolated from gills (5%) and viscera (1.66%). Shiga toxigenic E. coli (STEC) with genes eae + Stx2, Stx1 and Stx2 were isolated in viscera (5%) and gills (3.33%). For Enteroinvasive E. coli (EIEC), ial gene was isolated in gills (3.33%) but no ipah gene. Enterotoxinogen E. coli (ETEC) with lt gene and Enteroaggregating E. coli (EAEC) with aggA gene were not detected in this study. This study revealed that some fish from Layo farm are carriers of virulent E. coli that can cause serious human diseases and can lead to consumer death if cooking is insufficient or by cross-contamination. This therefore poses a real public health problem.
Key words: Escherichia coli pathogens, fish, ponds, public health.
Fish is one of the main sources of animal protein in human diet. In Côte d’Ivoire, production of fish ranges from 50.000 to 80.000 tons annually. This production covers 30% of the needs which went from 300 000 tons in 2005 to 850 000 tons in 2010. The deficit is filled by fish imports which represent 268 333 tons (67%) of national need for halieutic products (Coulibaly, 2010). To solve deficit problems, overfishing and extinction of some species, several aquaculture stations have emerged, including Layo. Layo aquaculture station located in Dabou department is an experimental site. Some ponds are fed directly by Ebrié lagoon or groundwater. Several studies have shown that waters of Ebrié lagoon are polluted because of human activities taking place around them (Kouassi et al., 1990; Adingra, 2007; Tuo et al., 2013). Recent work on waters in ponds of this experimental farm have revealed their strong contaminations with fecal coliforms and Vibrio (Toulé et al., 2017). But there is not enough information about sanitary quality of fish in these aquaculture sites. Relative information are about physico-chemical parameters and microbiological qualities of water. Also, this study was conducted to investigate the presence of Escherichia coli (indicator of faecal contamination) in fish Oreochromis niloticus in ponds at the Experimental Fish Farm (Layo) and to detect by PCR method, five pathogroups of E. coli like Enteropathogenic E. coli (EPEC), Shiga toxigenic E. coli (STEC), Enteroinvasive E. coli (EIEC), Enterotoxinogen E. coli (ETEC) and Enteroaggregating E. coli (EAEC) causing diarrhea, haemolytic uremic syndrome in human.
Sampling sites
This study was conducted in fish farm Layo. This site was located between 05°13’50,9’’ N and 004°26’25,1’’ W, contained 18 ponds whose sizes varied from 200 to 900 m2, 0.70 to 1.30 m of water depth and fed by Ebrié lagoon and groundwater. Four ponds, E5, E6, E11 and E13 were selected according to water sources (Table 1).
Water quality sampling and measurement
Water quality parameters like, pH, salinity, temperature, and dissolved oxygen were measured in situ using a multiparameter YSI 6920 V2-1S (USA). Water samples were taken approximately at 20 cm of depth using 1000 mL borosilicate bottles and stored in a cooler containing ice before transported to laboratory within 4 h. The determination of suspended solids (SS) was made according to the centrifugation method (Rodier et al., 2009). Nitrate (NO3-) was measured by the cadmium reduction method (HACH method 8039) and the nitrite method (NO2-) by the diazotisation method (HACH method 8507).
Fish sampling
Sampling was performed in April 2015. In four ponds selected, five fish of O. niloticus were taken per ponds, placed in individual labeled sterile polypropylene plastic bags, kept on ice and transported to laboratory within 4 h. Their body weight ranged from 300 to 500 g, and their length between 20 and 30 cm. In total, 20 gills and 20 viscera were analyzed.
Isolation and identification of E. coli in viscera and gills of fish
For analysis, 25 g of viscera and gills from each sample were added to 225 ml of sterile buffer peptone water contained in a sterile plastic stomacher bag and mixed well and incubated at 44°C for 24 h. After, 0.1 mL of solution was inoculated on desoxycholate agar (Becton Dickinson, GmbH) and Petri dishes were incubated at 44°C for 24 h. Red colonies were used as presumptive E. coli. Three colonies of E. coli per Petri dishes were purified and confirmed by positive indole, negative citrate and urea. E. coli strain of American Type Culture Collection 25922 (ATCC 25922) was used as control.
Detection of virulence genes by PCR
Detection of virulence genes (Table 2) were made in 120 strains of E. coli from viscera (60 strains) and gills (60 strains). DNA of each isolate was extracted according to boiling method. Three colonies of an overnight bacterial culture were taken and suspended in 500 ðœ‡L of distilled water. DNA was purified according the modified method described by Ausubel et al. (1992). The mixture was stored at -20°C for 10 min and then boiled at 100°C for 10 min. 400 μl of phenol/chloroform mixture (24:1) were added to 150 μl of supernatant. The tube was vortexed for 2 min and centrifuged at 13000 rpm for 2 min. Top phase was recovered into a new tube and the lower phase was discarded. On the upper phase obtained, 1/10th volume of sodium acetate to 3 M and 200 μl of absolute ethanol were added and stored at -20°C for precipitation and the mixture was incubated for 1 h at -20°C. After incubation, they were centrifuged again at 13000 rpm for 20 min at +4°C and then supernatant is removed by flipping the tubes. 1 ml of 70% ethanol stored at -20°C was added to the pellet and tubes were centrifuged again at 13000 rpm for 5 min. Then the supernatant was removed by flipping the tubes. The pellet was dried at the heating block for 15 min at 95°C and 100 μL of nuclease-free water were added to each tube.
According to modified methods of multiplex PCR previously described by Dadie et al. (2014), eight genes were screened in this study.
Multiplex PCR for Stx1, Stx2 and lt genes detection
The PCR was performed in a final volume reaction of 25 μL containing 8.25 μL nuclease-free water (Ambion), 5 μL PCR buffer (5X), 1.5 μL magnesium chloride (MgCl2, 25 mM) (Promega Corporation, Madison, USA), 0.5 μL Deoxynucleotide Triphosphates (dNTPs, 10 mM), 0.75 μL of each primer (20 mM) (Table 2), 0.25 μL Go Tag®G2 Flexi DNA polymerase 5 U/μL (Promega Corporation, Madison, USA) and 5 μL of template DNA.
For positive control of gene, E. coli strains previously known were used (Dadie et al., 2014). The amplification program used for Stx1, Stx2 and lt genes, included an initial denaturation (94°C, 3 min), followed by 35 cycles each composed of initial denaturation (94°C, 30 s), primer annealing (57°C, 45 s) and extension (72°C, 30 s) and a final extension (72°C, 30 s). After these cycles, a final extension of 5 min at 72°C is realised.
Multiplex PCR for invasive and adhesine genes (eae, bfpA, ial, aggA, ipah)
This second PCR was performed in a final volume reaction of 25 μL containing 5.25 μL nuclease-free water (Ambion), 5 μL PCR buffer (5X), 1.5 μL magnesium chloride (MgCl2, 25 mM) (Promega Corporation, Madison, USA), 0.5 μL Deoxynucleotide Triphosphates (dNTPs, 10 mM), 0.75 μL of each primer (20 mM) (Table 2), 0.25 μL Go Tag®G2 Flexi DNA polymerase 5 U/μL (Promega Corporation, Madison, USA) and 5 μL of template DNA.
For invasive and adhesine genes (eae, bfpA, ial, aggA, ipah), the program was an initial denaturation step at 94°C for 3 min, followed by 35 cycles of denaturation (94°C for 30 s), primer annealing (56°C for 20 s), and extension (72°C for 30 s), with a final extension at 72°C for 5 min. Primers used in these PCR were reported in Table 2. PCR amplification products were revealed on a gel Doc EZ® imager (Bio-Rad) after electrophoresis in 2% agarose gel containing Syber safe (Invitrogen).
Physico-chemical parameters in ponds
Physico-chemical results are presented in Figure 1. Water temperature in four selected ponds ranged from 35 to 36°C. This variability may be the fact that Côte d'Ivoire has a tropical climate (Inza et al., 2009) and these high temperatures could be explained by solar radiation according to Lwamba et al. (2015). High temperature is also an important predisposing factor for the growth of harmful bacteria for fish in aquaculture (Zhang et al., 2016). Salinity in all ponds was low and ranged from 0.25 (E11) to 0.650/00 (E5) certainly because they are fed by inland waters and Ebrié lagoon is under influence of Agneby River. Agneby River after having crossed its watershed, discharges its effluents in Ebrie lagoon (Toulé et al., 2017) and is the seat of a strong anthropic pressure (Kamagate et al., 2017). Suspended solids (SS) ranged from 44.2 (E13) to 57.2 mg L-1 (E6). These values are higher than those observed (7.14 mg L-1) in floating cages in Ebrié lagoon at Jacqueville aquaculture station (Toulé et al., 2017). These high values in Layo ponds could be explained by the fact that they are closed ecosystems where water renewal is rare. In rearing structures, fish are fed by artificial food. As a result, SS could be attributed to the enormous amounts of organic matter produced from uneaten food and fish metabolite waste. The resuspension of SS during aquaculture activities (sexing, fishing, sorting, transfer, etc.), rain penetration and runoff of soil and plant particles to ponds and pens could result in increased levels in SS.
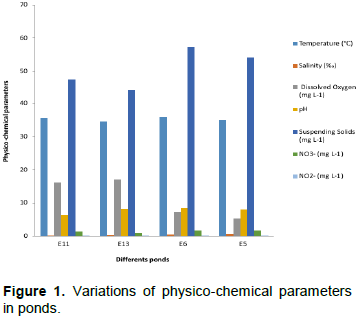
The smallest dissolved oxygen values were identified in ponds E5 (5.32 mg L-1) and E6 (7.43 mg L-1), while the highest values were found in ponds E11 (16.19 mg L-1) and E13 (17.14 mg L-1). The higher dissolved oxygen in pond E13 where suspended matter is the lowest could be due to the fact that it is directly under the influence of Ebrié lagoon. pH varies from 6.29 (E11) to 8.5 (E6).
Virulence gene in fish
Frequency of E. coli strains isolated
E. coli was detected (100%) in all fish analyzed (20 fish). 120 E. coli isolates were detected through biochemical identification. Sixty came from gills and 60 from viscera of O. niloticus obtained from Layo ponds. E. coli is a normal habitant of warm-blooded animals. Although not a normal host of fish flora, it has been isolated from stomach, gut, gills, muscle, and skin of fish (Janssen, 1970; Hejkel et al., 1983; Ogbondeminu, 1993; Kouadio-Ngbesso et al., 2016; Ribeiro et al., 2016). The presence of E. coli in viscera and gills in Layo's fish may be due to the quality of water in which they are cultured. Fish are intermediate carriers of these microorganisms. Indeed, some works revealed the presence of heavy loads faecal coliforms and Vibrio in the ponds of Layo (Toulé et al., 2017). Research conducted by Koteswar et al. (2017), on 22 finfish from ponds in aquaculture center of Mangalore in India, revealed E. coli in 86.6% of samples. Ponds are closed environments where horizontal circulation and vertical trade are slowed down, feed intake and releases of captive fish metabolites accumulate in the sediments, providing a good source of nutrients for bacteria. Previous work highlighted penetration and installation of bacteria in different tissues and organs of fish living in a polluted environment (Gariboglio et al., 1976; Pal and Dasgupta, 1992).
Virulence genes detected among E. coli isolated
The results of the genetic analysis of E. coli isolates from fish samples are presented in Table 3. Of the 120 E. coli isolates, 11 strains (9.16%) were positive for virulence genes. In gills, the pathogenic strains of E. coli were more isolated with a frequency of 11.66% (7 strains) than in viscera with 6.66% (4 strains). EPEC represented by eae gene were identified in 5% (3 strains) and in 1.66% (1 strain) in gills and viscera strains, respectively. The lack of bfpA gene in the present study suggests that EPEC strains are probably atypical. This kind of EPEC (eae +, bfpA-, Stx-) was isolated from acute diarrhea (Vieira et al., 2001), and would be predominant in strains from developed countries (Paciorek, 2002; Trabulsi et al., 1996). EPEC is the leading cause of childhood diarrhea in developing countries (Food and Drug Administration, 2012). It damages the epithelial cells of the small intestine by producing typical lesions (Kaper et al., 2004). Shigatoxigenic gene Stx1 was identified only in viscera for 1.66% (1 strain) while Stx2 was identified in viscera at 3.33% (2 strains) and in gills at 3.33% (2 strains). The positive isolate for both Stx2 and eae, characteristic of Enterohemoragique E. coli (EHEC) but included in STEC, was identified in gills at 1.66% and in viscera at 1.66% in 1 strain. The presence of Shigatoxigenic genes has also been identified in intestines of fish from ponds in northeast of Sao Paulo, Brazil (Ribeiro et al., 2016). The presence of these genes is predominant in patients with Hemolytic Uremic Syndrome. Although the detection of strains carrying both eae and Stx genes in aquaculture environments is low (Zschock et al., 2000; Irino et al., 2005; Alagarsamy et al., 2009), it has been detected in the present work and in approximately 7.69% of intestinal fish strains from ponds in northeast of Sao Paulo (Ribeiro et al., 2016). Invasive gene ial was identified only in gills (3.33%) but ipah invasive gene was not detected in othe present study. Regarding EIEC, previous studies revealed the rarity of this pathotype in environment or in food (Barbosa et al., 2014). It is more often isolated in fish from developing countries (Peng et al., 2009). This pathotype is also responsible for infantile diarrhea (Moreno et al., 2010; Nguyen et al., 2005). The genes encoding ETEC (lt, st) and EAEC (agg A) were not identified in this study (Table 3).
All pathogenic genes identified in this study are likely to cause diarrhea, dysentery, haemorrhagic colitis, haemolytic uremic syndrome and chronic post-infection sequelae in men (Ribeiro et al., 2016). Their detections in gills and viscera of fish could come from direct contact with water (Fouz et al., 2000). These results are also identified in the viscera of O. niloticus from aquaculture stations in Sao Paulo, Brazil (Ribeiro et al., 2016). In contrast to this work, pathogenic strains were not detected in fish caught in Aby Lagoon, Côte d'Ivoire by Kambire et al. (2017).
The majority (80%) of the genes sought, Stx2, eae, ial, and eae + Stx2, were detected in fish strains from E6 pond. The Stx2 gene was detected in E5, E6 and E11 ponds. The ial gene was present in strains from E13 and E6 ponds. At least 1 gene was detected per pond (Figure 2).
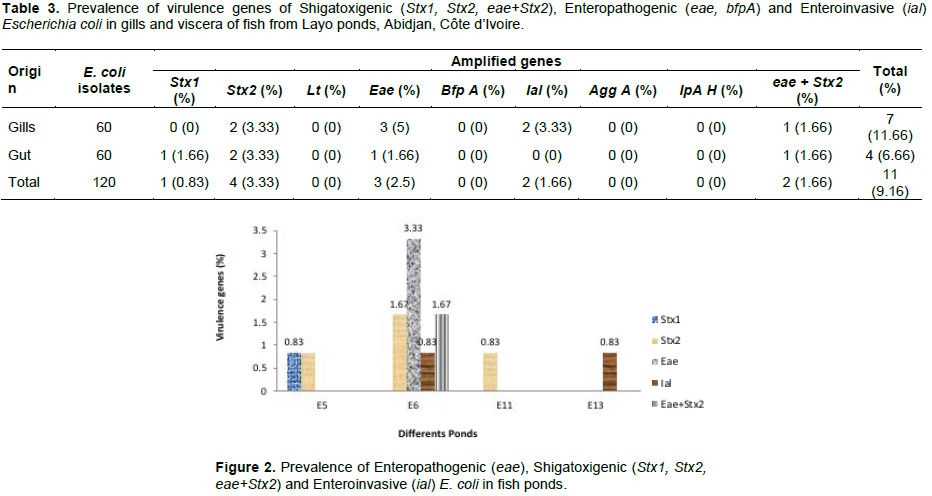
The aquaculture station of Layo is close to a village where there is no septic system and animals such as chicken, sheep, goats and other domesticated animals move around freely. Bovine faeces have been identified as the main reservoir of E. coli and are a vehicle of transmission to the environment, cattle and food (Wang et al., 1996). Environmental conditions surrounding crop sites may affect the quality of water and farmed fish. The prevalence of these pathotypes reflects the bacterial compositions of the living environments (water and sediments) and the health status of the fish according to Pakingking et al. (2015). In aquaculture activities, few studies have been conducted on the presence of microorganisms responsible for human diarrhea. In addition, the presence of E. coli in environment is generally quantitatively assessed as an indicator of faecal contamination with respect to the quality of irrigation water, without considering that the presence of this microorganism, even at low concentration, indicates a risk of transmission of pathogens to humans. It is true that E. coli bacteria do not cause losses in aquaculture production, but it can cause human diseases. Therefore, fish farmers do not see the need to apply appropriate health control measures to ensure product quality. However, infected fish used as a food source can serve as means of transmission of these agents to humans, and even contaminate other surfaces.
This study revealed the presence of E. coli pathotypes like STEC, EPEC, EIEC in gills and viscera of O. niloticus in ponds of Layo fish farm. However, ETEC and EAEC pathotypes were not detected. Although these pathogenic bacteria do not cause losses in fish production, they cause serious human diseases that can lead to death. There is therefore a real public health problem that should be of concern and brought to the attention of the appropriate government authorities.
The authors have not declared any conflict of interests.
REFERENCES
|
Adingra AA (2007). Organic and bacterial pollution of waters in Côte d'Ivoire: case of a rural area (experimental aquaculture site Layo) and urban area Ebrié lagoon. PhD thesis, University of Cocody, Abidjan (Côte d'Ivoire), 184 p.
|
|
|
|
Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K (1992). Current protocols in Molecular Biology. New York: Greene Publishing Association; Wiley-Interscience.
|
|
|
|
|
Alagarsamy S, Thampuran N, Joseph T (2009). Virulence genes, serobiotypes and antibiotic resistance profile of Escherichia coli strains isolated from aquaculture and other sources. Aquaculture Research 41:1003-1014.
Crossref
|
|
|
|
|
Barbosa MM, Pinto FD, Ribeiro LF, Guriz CS, Ferraudo AS, Maluta RP, Rigobelo EC, Ávila FA, Amaral LA (2014). Serology and patterns of antimicrobial susceptibility in Escherichia coli isolates from pay-to-fish ponds. Arquivos do Institudo Biológico., Sao Paulo 81(1):43-48.
Crossref
|
|
|
|
|
Coulibaly R (2010). Analyse of fishing contribution to Ivorian economy. University of Cocody-Abidjan DESS, High Studies in Management of Economic Policy Auditor GPE, 11th promotion 30 p.
|
|
|
|
|
Dadie A, Kouassi N, Dako E, Dje M, Dosso M (2014). Virulence, serotype and phylogenetic groups of diarrhoeagenic Escherichia coli isolated during digestive infections in Abidjan, Côte d'Ivoire. African Journal of Biotechnology 13(9):998-1008.
Crossref
|
|
|
|
|
Food and Drug Administration (2012). The bad bug books. 2nd edn, 292 p.
|
|
|
|
|
Fouz B, Toranzo AE, Milan M, Amaro C (2000). Evidence that water transmits the disease caused by the fish pathogen Photobacterium damselae subsp. damselae. Journal of Applied Microbiology 88:531-535.
Crossref
|
|
|
|
|
Frankel G, Giron JA, Valmassoi J, Schoolnik GK (1989). Multi-gene amplification: simultaneous detection of three virulence genes in diarrhoeal stool. Molecular Microbiology 3:1729-1734.
Crossref
|
|
|
|
|
Gariboglio M, Ebbeke E, Merlassino M (1976). Fecal bacteria indicators in the intestinal content of fresh water ï¬sh. Limnobios 1:9-100.
|
|
|
|
|
Hejkel TW, Gerba CP, Henderson S, Freeze M (1983). Bacteriological, virological and chemical evaluation of a wastewater aquaculture system. Water Research 17:1749-1755.
Crossref
|
|
|
|
|
Inza B, Métongo BS, Assoi OE, Albert T, Yobou B (2009). Physicochemical characterization of water and surface sediments in the bay of billionaires, Ebrié Lagoon, Côte d'Ivoire. Ivorian Review of Science and Technology 13:135-154.
|
|
|
|
|
Irino K, Kato MA, Vaz TM, Ramos II, Souza MA, Cruz AS, Gomes TA, Vieira MA, Guth BE (2005). Serotypes and virulence markers of Shiga toxin producing Escherichia coli (STEC) isolated from dairy cattle in Sao Paulo State, Brazil. Veterinary Microbiology 105:29-36.
Crossref
|
|
|
|
|
Janssen WA (1970). Fish as potential vectors of human bacterial diseases of ï¬shes and shellï¬shes. American Fisheries Society Special Publications 5:284-290.
|
|
|
|
|
Kamagate B, Dao A, Noufe D, Yao KL, Fadika V, Gone DL, Savane I (2017). Contribution of GR4J model for modeling Agneby watershed runoff in SouthEast of Côte d'Ivoire. Larhyss Journal (29):187-208.
|
|
|
|
|
Kambiré O, Adingra AA, Yao KM, Nevry-Koffi R (2017). Prevalence of virulence genes associated with diarrheagenic pathotypes of Escherichia coli isolates from water, sediment, fish, and crab in Aby Lagoon, CoÌ‚te d'Ivoire. International Journal of Microbiology 2017:1-8.
Crossref
|
|
|
|
|
Kaper JB, Nataro JD, Mobley HLT (2004). Pathogenic Escherichia coli. Nature Reviews Microbiology 2:123-139.
Crossref
|
|
|
|
|
Koteswar BA, Devivaraprasad R, Ravi G, Karunasagar I (2017). Occurrence of pathotypes of Escherichia coli in aquatic environment. International Journal of Current Microbiology Applied Sciences 6(9):3266-3275.
Crossref
|
|
|
|
|
Kouadio-Ngbesso N, Kouassi N, Kouadio F, Adingra A, Yobouet BA, DadieÌ A, Dje KM (2016). Relationship between the phylogenetic group of Escherichia coli strains isolated in water and fish in Fresco lagoon (CoÌ‚te d'Ivoire). International Journal of Current Microbiology Applied Sciences 5(10):413-423.
Crossref
|
|
|
|
|
Kouassi AM, Guiral D, Dosso M (1990). Seasonal variations of microbial contamination of urban area of a tropical estuarine lagoon case of Abidjan city (Côte d'Ivoire). Tropical Hydrobiology Journal 23(3):181-194.
|
|
|
|
|
Luscher D, Athwegg M (1994). Detection of shigellae, enteroinvasive and enterotoxigenic Escherichia coli using the polymerase chain reaction (PCR) in patients returning from tropical countries. Molecular and Cellular Probes 8:285-290.
Crossref
|
|
|
|
|
Lwamba BJ, Mama K, Kiwaya AT, Ipungu LR, Nyongombe UN (2015). Variations of water temperature in pond during the cold period in Lubumbashi (Congo Democratic Republic) and implications for fish production. Journal of Applied Biosciences 90:8429-8437.
Crossref
|
|
|
|
|
Moreno ACR, Fernandes-FIilho A, Gomes TAT, Ramos STS, Montemor LPG, Tavares VC, Santos Filho L, Irino K, Martinez MB (2010). Etiology of childhood diarrhea in the northeast of Brazil: significant emergent diarrheal pathogens. Diagnostic Microbiology and Infectious Disease 66(1):50-57.
Crossref
|
|
|
|
|
Nguyen TV, Le PV, Le CH, Weintraub A (2005). Antibiotic resistance in diarrheagenic Escherichia coli and Shigella strains isolated from children in Hanoi, Vietnam. Antimicrobial Agents Chemotherapy 49(2):816-819.
Crossref
|
|
|
|
|
Ogbondeminu FS (1993). The occurrence and distribution of enteric bacteria in fish and water of tropical aquaculture ponds in Nigeria. Journal of Aquaculture in the Tropics 8:61-66.
|
|
|
|
|
Paciorek J (2002). Virulence properties of Escherichia coli faecal strains isolated in Poland from healthy children and strains belonging to serogroups O18, O26, O44, O126 and O127 isolated from children with diarrhoea. Journal of Medical Microbiology 51:548- 556.
Crossref
|
|
|
|
|
Pakingking RJr, Palma P, Usero R (2015). Quantitative and qualitative analyses of the bacterial microbiota of tilapia (Oreochromis niloticus) cultured in earthen ponds in the Philippines. World Journal of Microbiology and Biotechnology 31(2):265-275.
Crossref
|
|
|
|
|
Pal D, Dasgupta Ch (1992). Microbial pollution in water and its effect on ï¬sh. Journal of Aquatic Animal Health 4:32-39.
Crossref
|
|
|
|
|
Peng J, Yang J, QI J (2009). The molecular evolutionary history of Shigella spp. and enteroinvasive Escherichia coli. Infection, Genetics and Evolution 9(1):147-152.
Crossref
|
|
|
|
|
Ribeiro LF, Barbosa MMC, Pinto F, Guariz CSL, Maluta RP, Rossi JR, Rossi GAM, Lemos MVF, Amaral LA (2016). Shiga toxigenic and enteropathogenic Escherichia coli in water and fish from pay-to-fish ponds. Letters in Applied Microbiology 62:216-220.
Crossref
|
|
|
|
|
Rodier J, Legube B, Merlet N (2009). Water analyse. 9th edition. Dunod: Paris 1579 p.
|
|
|
|
|
Schmidt H, Beutin L, Karch H (1995). Molecular analysis of the plasmid encoded hemolysin of Escherichia coli O157:H7 strain EDL 933. Infection and Immunity 63:1055-1061.
|
|
|
|
|
Svenungsson B, Lagergren A, Ekwall E, Evengard B, Hedlund K, Anders KA, Lofdahl S, Lennart SL, Weintraub A (2000). Enteropathogens in Adult Patients with Diarrhea and Healthy Control Subjects: A 1-Year Prospective Study in a Swedish Clinic for Infectious Diseases. Clinical Infectious Diseases 30 (5): 770-778.
Crossref
|
|
|
|
|
Toulé AC, Adingra A, Kouadio-N'gbesso N, Kambire O, Koffi-Nevry R, Koussemon M (2017). Physicochemical and bacteriological characterization of waters in Layo and Jacqueville aquaculture
|
|
|
|
|
stations (Ebrié lagoon, Côte d'Ivoire). International Journal of Biological and Chemical Sciences 11(6):2842-2855.
|
|
|
|
|
Trabulsi L, Campos L, Whittam T, Gomes T, Rodrigues J, Goncalves A (1996). Traditonal and non-traditional enteropathogenic Escherichia coli serogroups. Revista de Microbiologia 84:585-592.
|
|
|
|
|
Tuo AD, Yeo KM, Soro MB, Trokourey A, Bokra Y (2013). Contamination by nutrients and heavy metals in Ebrie Lagoon (Abidjan, Ivory Coast). International Journal of Chemical Technology 6:198-209.
Crossref
|
|
|
|
|
Vieira RHSF, Rodrigues DP, Gocalves FA, Menezes FGR, Aragao JS, Sousa OV (2001). Microbicidial effect of medicinal plant extracts (Psidium guajava Linn. and Caricapapaya Linn.) upon bacteria isolated from fish muscle and known to induce diarrhea in children. Revista Do Instituto De Medicina Tropical De Sao Paulo 43:145-148.
Crossref
|
|
|
|
|
Wang G, Zhao T, Doyle MP (1996). Fate of enterohemorrhagic Escherichia coli O157:H7 in bovine feces. Applied and Environmental Microbiology 62:2567-2570.
|
|
|
|
|
Yatsuyanagi J, Saito S, Saito H, Miyagima Y, Amano K, Enomoto K (2002). Characterisation enteropathogenic and enteroaggregative Escherichia coli isolate from diarrhoeal outbreaks. Journal of Clinical Microbiology 40(1):294-297.
Crossref
|
|
|
|
|
Zhang D, Xu D, Shoemaker C (2016). Experimental induction of motile Aeromonas septicemia in channel catfish (Ictalurus punctatus) by water- borne challenge with virulent Aeromonas hydrophila. Aquaculture Reports 3:18-23.
Crossref
|
|
|
|
|
Zschock M, Hamann H, Kloppert B, Wolter W (2000). Shiga toxin producing Escherichia coli in faeces of healthy dary cows, sheep and goats: prevalence and virulence properties. Letters in Applied Microbiology 31:203-208.
Crossref
|
|