The king weakfish (Macrodon ancylodon) has a wide geographic distribution in South America and provides a positive economic outlook for the fishing industry in Brazil. The present study aimed to identify the intestinal microbiota and bacterial strains that can increase during storage of the king weakfish. Characterization of the microbiological quality was performed on samples to identify the microflora through isolation on culture media and identification using BD BBL kits for Gram-negative and Gram-positive bacteria. The growth of isolated strains was tested at conventional storage temperatures of 7 and 3°C used to limit the expansion of pathogenic bacteria populations. All king weakfish samples were within the limits established by Brazilian law for microbiological standards. Twenty strains that were isolated from the intestinal microbiota of M. ancylodon were identified; five stood out and may play a role in the safety and/or shelf-life of this fish species: Staphylococcus cohnii subsp. urealyticum, Burkholderia cepacia, Pseudomonas stutzeri, Pseudomonas fluorescens and Corynebacterium sp. However, only the first two strains showed significant growth at 3°C after three days of storage, mainly B. cepacia which is also considered an opportunistic pathogen.
The king weakfish (Macrodon ancylodon) is an important species nationally and along the northern coast of Brazil, in terms of both the volume captured and its consumption by the local population. The species M. ancylodon belongs to the family Sciaenidae and has a wide geographic distribution in South America, ranging from the tropical waters of Venezuela to the subtropical waters of Argentina. It can grow to an average of 40 cm long, weighing less than 1 kg, by feeding on shrimp, fish and squid (Santos et al., 2003).
Fish can harbor a large number of microorganisms that are pathogenic to humans and are acquired from environmental pollution as well as improper handling between capture and final consumption. Due to its high perishability, fish should be stored at low temperatures and handled under adequate sanitary conditions throughout the production chain in order to deliver a safe product to consumers (Mol and Tosun, 2011). Importantly, most pathogenic bacteria do not grow at temperatures below 4.4°C (Jay, 2005).
Improper handling results in significant post-capture losses of wild-caught and cultured fish, mainly due to microorganisms. Because they are extremely perishable, fish lose their initial freshness due to enzymatic and chemical reactions, which is closely followed by a complete loss in quality due to microbial activity that occurs primarily on the skin, gills and intestines (Gram and Dalgaard, 2002). During fish processing, microorganisms that are naturally found in the intestine and on the skin may contaminate the muscle, utensils and equipment, leading to cross-contamination and the transmission of food-borne illnesses due to improper handling and inadequate storage (Jay, 2005). It is important to identify the microbial species that contribute to spoilage and are involved in quality loss in fish such that appropriate quality control measures can be implemented.
The total number of microorganisms in the gastrointestinal tract of fish is small compared to that of warm-blooded animals and varies with the age, food source and environmental conditions of the fish (Nayak, 2010). The primary microbial groups that colonize the intestinal tracts of temperate water fish vary based on the fish species but overall are predominantly psychrotrophic, aerobic, anaerobic or facultative Gram-negative bacteria, including Pseudomonas, Moraxella, Acinetobacter, Shewanella, Flavobacterium, Cytophaga, Vibrio, Corynebacterium, Photobacterium and Alteromonas (Gram and Huss, 1996; International Commission on Microbiological Specifications for Foods (ICMSF), 2005). Some Gram-positive bacteria, including lactic acid-producing bacteria, Bacillus sp. and Staphylococcus sp., are also present (Lalitha and Surendran, 2006). However, only a few members of the fish microbiota, known as "spoilage-specific organisms" or SSOs, are responsible for spoilage (Gram and Dalgaard, 2002).
There are no current data available in the literature on the intestinal microbiota and there are few studies on the microbiological quality of the king weakfish (Macrodon ancylodon). Therefore, the aim of the present study was to identify the bacterial populations in the microflora, microbiological characterization this species and study the growth temperature characteristics of the micro-organisms during storage.
Samples
King weakfish were captured at sea, 70 km from the municipality of Salinópolis, located in the state of Pará, Brazil, and whole fish samples were collected directly from commercial fishing vessels after docking. Four fish were collected at two different periods (beginning and end) of the months of May and June, 2014, which are the months of highest annual production of the specie. The samples were placed in polyethylene bags and transported in coolers on ice to the Food Microbiology Laboratory, Federal University of Pará where the analyses were performed.
Microbiological quality of the samples
Twenty-five grams of each fish sample (ventral part of the fillet) were aseptically removed for each collection period. Samples were transferred to sterile pouches, and 225 ml of sterile saline peptone water was added. Samples were then homogenized at 2,300 rpm for 30 s (Stomacher 400 Circulator SEWARD). The total counts of heterotrophic aerobic mesophilic and aerobic psychrotrophic bacteria counts were determined using standard count in plates of plate count agar followed by incubation at 36°C for 48 h and 7°C for 10 days. Analyses of coliforms at 45°C, Salmonella sp. and coagulase-positive Staphylococci were also performed. All analyses were conducted according to the methods described in Compendium of Methods for the Microbiological Examination of Foods (Downes and Ito, 2001).
Intestinal microbiota
Bacterial isolation
Sample material was extracted from fecal matter and the internal intestinal walls of the king weakfish using a sterile loop. All extracted material was then homogenized in 10 ml of sterile saline peptone water. The entire procedure was performed using aseptic techniques. For identification of intestinal microbiota of the king weakfish the homogenates were inoculated in aliquots of 0.1ml on the surface using two types of agar media, violet red bile glucose (VRBG) agar for enterobacteria strains and Baird-Parker agar for gram-positive bacteria, and both plates were incubated at 36°C for 48 h. Next, one plate was selected for each medium and five to eight colonies per plate were randomly chosen and the selected colonies were striated to obtain a pure culture isolation on the same medium (VRBG or Baird-Parker Agar). After incubation at 36°C for 48 h, the colonies were transferred to tubes containing brain heart infusion broth (BHI) supplemented with 10% glycerol and stored frozen for subsequent identification.
Identification of bacterial strains
The isolated colonies were characterized using Gram stains for morphology (microscopy) as well as the oxidase and indole reactions. The strains were then identified using kit recognized worldwide of the BD BBL Crystal including the Enteric/Nonfermenter ID System for Gram-negative bacteria and the Gram-positive ID System for Gram-positive bacteria. The assays were performed according to the manufacturer's protocol.
Temperature limits for growth of the microflora
The methodology employed here was adapted from the method of Damasceno et al. (2015), where the optical density was used to identify strains that could grow at the different storage temperatures. The bacterial strains previously identified and frozen in BHI broth were first reactivated in proportion 1:10 ml in the medium for 24 h at 36°C. The 0.6 ml from isolates identified were then transferred to tubes containing 9 ml of BHI medium and incubated at 7°C (mean temperature for conventional refrigeration) and 3°C (below the temperature limits for the growth of pathogens) for 56 and 80 h, respectively. The spectrophotometric readings were taken at four-hour intervals using a UV spectrophotometer (Modelo Nova 2000) at different wavelengths determined based on the highest absorbance for each strain identified after scanning the entire spectrum in the UV-visible (200 to 800 nm).
Statistical analysis
Analysis of variance (ANOVA) and Tukey's test at a 5% significance level were used to determine the significance of the microbiological quality between samples of periods and the temperature limits for growth of the identified microorganisms. The program Statistica 8.0 was used for these analyses.
Characterization of the microbiological quality
All samples were within the microbiological standards established by the Brazilian legislation for fresh fish, demonstrating the good quality of the samples collected at the four time periods in May and June. This standard was based on the absence of Salmonella spp. and less than 103 coagulase-positive Staphylococci in 25-gram samples of king weakfish (Brazil, 2001). The results of the additional analyses used to characterize the microbiological quality of the samples are presented in Table 1.

Total and thermotolerant coliforms are used as quality indicators because fish do not usually carry those organisms, particularly E. coli and faecal coliform, their presence reflect the degree of microbial contamination to which food is exposed (ICMSF, 1986). The results of the analysis of coliforms at 45°C indicated low levels of contamination for the king weakfish samples over the collections periods. In the standard plate count analysis for aerobic mesophiles and psychrotrophs, all samples were below the total counts established by the International Commission on Microbiological Specifi-cations for Foods (ICMSF, 2005), which recommends a maximum of 7 Log CFU/g of refrigerated fish. Counts of psychrotrophic bacteria were higher than those reported by Thong et al. (2013) for the species Pangasius hypophthalmus at 4.3 Log CFU/g. In addition, the total counts for these microorganisms were higher than the counts for mesophiles, indicating that the storage of king weakfish until 4°C favors the growth of microorganisms spoilage (Jay, 2005).
Identification of the intestinal microbiota
Table 2 lists the bacteria identified from the intestinal microbiota of king weakfish. Twenty isolates were evaluated using bacterial identification kits; 90% (18 strains) were Gram-negative and 10% (two strains) were Gram-positive. However, five isolates could not be identified using the kit standards or the manufacturer's identification software. There is ample evidence that the gastrointestinal microbiota of fish is highly variable, largely reflecting the aquatic environment and food source of each fish (Al-Harbi and Uddin, 2004). The microbiota of most fish is dominated by psychrotrophic Gram-negative species; however, the bacterial load in tropical fish often contains more Gram-positive and enteric bacteria than temperate fish species (Gram and Huss, 1996). Gram-positive strains such as Staphylococcus spp., Micrococcus spp. and Bacillus spp. have been isolated from tropical marine fish, although reports on the identity or specific sources of these bacteria are limited (Al-Bulushi et al., 2010).

Most bacteria identified by the technique used have a very high similarity profile (over 90%), demonstrating security in the identification of the microorganism in microflora. In general, the identified strains from the king weakfish intestine are considered spoilage bacteria and may directly play a role in the safety and/or shelf-life of this fish species. Staphylococci are not part of the normal fish microbiota (Van den Broek et al., 1984), given that coagulase-negative Staphylococci are normally found in the mucous membranes and skin of samples of human origin (Piette and Verschraegen, 2009). The Staphylococcus cohnii subsp. Urealyticum isolated from king weakfish intestines was classified as a coagulase-negative Staphylococcus and is able to form large colonies and aerobically produce acid byproducts when metabolizing lactose (Tammy and Bannerman, 1996). Because it is not part of the normal fish microbiota, these bacteria only accounted for 6.67% of the strains identified in king weakfish. Most Staphylococcal species are harmless, although some species of this genus cause various diseases by producing enzymes and toxins, such as S. cohnii, which encodes a gene for enterotoxin production (Zell et al., 2008).
Studies characterizing the microbiota of foods in natura and food products are important in assessing the safety, spoilage and the influence of these microorganisms on sensory characteristics. Studies on salted and dried salted cod identified Gram-positive bacteria such as S. cohnii and Gram-negative bacteria such as Pseudomonas fluorescens, a bacterium that is associated with food spoilage due to its ablity to produce H2S and/or biogenic amines, especially in foods stored at low temperatures (Rodrigues et al., 2003). In the present study, 46.67% of the isolates from the king weakfish intestinal microbiota were identified as P. fluorescens (Table 2). P. fluorescens, an aquaculture pathogen that also infects many other fish species, produces extracellular enzymes that contribute to skin diseases, primarily in fish that are injured during improper handling and transport (Zhang et al., 2009). However, these and other virulence mechanisms of P. fluorescens remain poorly understood (Zhang et al., 2009).
Over the past decade, some non-fermentative Gram-negative bacteria have emerged as important fish and human pathogens (Enoch et al., 2007). In addition to the Gram-negative bacterium, this study identified two other strains Burkholderia cepacia and Pseudomonas stutzeri accounted for 26.67 and 13.33%, respectively, of the intestinal microbiota in king weakfish. B. cepacia is a well-known pathogen opportunistic associated with hospital-acquired infections (Enoch et al., 2007) due to its resistance to many antimicrobial agents (Spencer, 1995).
Miranda and Zelmeman (2002) reported antibiotic resistance in Gram-negative bacteria, which they attributed to the use of antibiotics to control bacterial pathogens in salmon farming. The authors evaluated strains isolated from water, food and minnows, where the most prevalent species was P. fluorescens. Other Gram-negative bacteria were also resistant, including Pseudomonas aeruginosa, S. maltophilia, B. cepacia and Acinetobacter. This factor has been extensively explained about the low permeability of the outer-membrane of these strains, which excludes a wide range of antimicrobial compounds (antibiotics, dyes and organic solvents) (Hancock, 1998).
Pseudomonas stutzeri is also known for its metabolic diversity; it is widely distributed in natural environments and frequently isolated from contaminated soil and wastewater (Lalucat et al., 2006). In a study that included more than 100 bacterial isolates from the intestinal tract of fish in Pakistan, the marine bacterium P. stutzeri isolated from the intestinal tract of ribbonfish (Desmodema spp.) exhibited antimicrobial activity against various bacterial species, including antibiotic resistant for Staphylococcus aureus (Lalucat et al., 2006). However, besides the ability to act as a producer of the antimicrobials, some microorganisms isolated from fish may be responsible for foodborne diseases. Al-Harbi and Uddin (2004) reported that the bacterial microbiota in hybrid tilapia intestines varied seasonally, and the species Aeromonas hydrophila, Shewanella putrefaciens, Corynebacterium urealyticum, Escherichia coli and Vibrio cholerae were the predominant bacterial isolates. Also isolated strains of opportunistic bacteria including Flavobacterium sp. Pseudomonas fluorescens, Salmonella sp., Staphylococcus sp. and Streptococcus sp., which have high importance among microorganism presents in intestinal microflora of fish, because under stress conditions, these bacteria may be food poisoning food agents (Al-Harbi and Uddin, 2004).
In addition to the seasonal factors that influence the bacterial microbiota, some storage conditions also alter bacterial colonization in fish. Lalitha and Surendran (2006) characterized the microorganisms in refrigerated shrimp, and during the first two weeks of storage, the microbiota was largely composed of the Gram-negative bacterial genera Aeromonas, Shewanella, Moraxella and Pseudomonas and the Gram-positive genera Enterococcus, Micrococcus and Corynebacterium. At the end of storage, the predominant organisms were Aeromonas, S. putrefaciens and Pseudomonas. Oku and Amakoromo (2013) also isolated 12 bacterial strains in freshwater fish in Nigeria, and the predominant strains were identified as B. subtilis, Corynebacterium, Lactobacillus, Pseudomonas and S. aureus. The identification of spoilage or pathogenic bacteria in the microbiota of fish species during their shelf life and an understanding of how they cause spoilage is extremely important in preserving fish quality under different storage conditions (temperature) and maintaining commercial stability.
Temperature growth limits of the microbial isolates
Monitoring the microbial content in the internal organs of fish helps estimate how their food-related qualities will be affected when stored under appropriate conditions during their storage. Therefore, the growth of bacterial isolates from the king weakfish microflora was assessed at 7 and 3°C, which correspond to conventional storage temperatures in household refrigerators and are growth-limiting conditions for most pathogenic bacteria. Figure 1 illustrates the growth of king weakfish intestinal microbiota isolates at 7°C for 56 h. None of the five strains showed significantly different (P > 0.05) growth rates over time. These data indicate that over a storage period of more than two days at a temperature of 7°C, neither Gram-negative or Gram-positive bacteria isolated from the intestine showed a significant increase in growth.
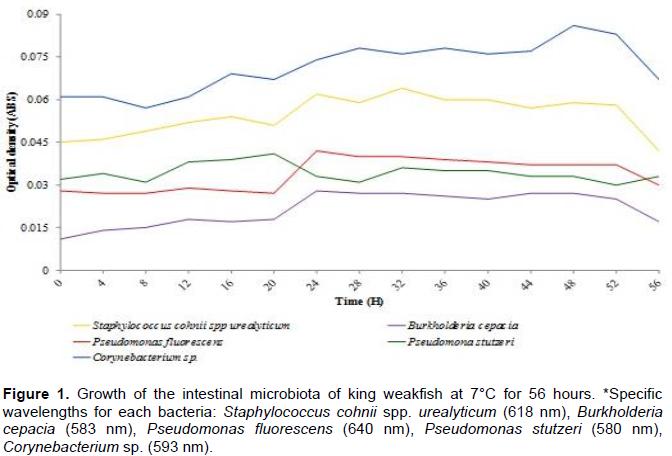
At a temperature of 3°C (Figure 2), the spoilage bacteria S. cohnii subsp. urealyticum and the opportunistic pathogenic B. cepacia, showed significant differences between their growth over time (P < 0.05). The growth of these microorganisms can be attributed to their ability to better adapt to the storage in low temperatures in experiment cultivation medium.

Damasceno et al. (2015) assessed the growth of bacterial isolates at 10 and 15°C for six hours and found that all 16 of the strains identified that composed the microbiota of the muscle of the butterfly peacock bass (Cichla ocellaris) and the piraiba catfish (Brachyplatystoma vaillantii) could not grow under the studied conditions. The authors further noted a correlation between high-temperature habitats (fish from tropical regions) and microbiological quality, decreases pathogens ability and limits the spoilage processes when these fish are stored at temperatures below 10°C. This behavior was also observed for king weakfish, as the identified isolates showed slow growth because of characteristics: good microbiological quality of samples fish and difficult in growth growth of some bacterial present in the intestinal microflora.
The acceptable microbiological quality and according to the identification of intestinal microbiota, the king weakfish can extend its shelf-life when stored at low temperatures. This affirmative is based on low counts of mesophilic, psychrotrophic bacteria and thermotolerant coliforms and the five species identified in microflora can cause deterioration and/or may play a role of opportunistic pathogen, but that did not grow significantly at 7°C after 56 h. However, at 3°C, the bacteria S. cohnii subsp. urealyticum and opportunistic pathogens B. cepacia showed significant growth, implying in the food safety after three days if stored under these conditions.
The authors have not declared any conflict of interest.



