Full Length Research Paper
ABSTRACT
“Rabilé” is a popular traditional ferment in Burkina Faso, consisting mainly of yeasts. It is used as a food supplement or additive like Single Cell Protein (SCP). The present work focused on identifying yeast microbiota in local food, and studying their growth kinetic parameters. “Rabilé” sampling from the 13 regions of Burkina Faso was used to isolate yeast strains. Molecular methods, including PCR-RFLP, Sanger Sequencing, and Single Locus Analysis, were applied for strain identification. The kinetic parameters were determined in batch culture. The results show 390 isolates belonging to 12 species with a predominance of Saccharomyces cerevisiae, followed by Cutaneotrichosporon curvatus. Among the selected strains, S. cerevisiae OG22 and Kluyveromyces marxianus KY01 showed the highest maximum growth rate (0.566 and 0.568 h-1) concerning kinetic parameters. These results demonstrate that “Rabilé” is an important biotope of yeast strains, and could be a potential food supplement.
Key words: Yeast, Rabilé, traditional ferment, molecular identification, kinetic parameters.
INTRODUCTION
Yeasts have played an essential role in the human diet for millennia (Bekatorou et al., 2006; Gómez-Pastor et al., 2011; Steensels and Verstrepen, 2014). They are used in brewing, bread-making, winemaking, and producing many other fermented foods worldwide (Bekatorou et al., 2006; Carrau et al., 2015). Nowadays, advances in genetic engineering (recombinant protein) have made it possible to extend their application in many fields such as the environment, the food industry, and health (Feldmann, 2012; Fallah et al., 2016; de Souza Varize et al., 2019).
In West Africa, yeasts are mainly used to produce ethyl alcohol and fermented foods (Somda et al., 2011; Tankoano et al., 2017; Johansen et al., 2019). The production of fermented foods is an income-generating activity for local producers (Lyumugabe et al., 2012; Djêgui et al., 2015), who provide enriched and inexpensive food. Yeasts improve the nutritional and organoleptic quality of these foods by providing additional proteins, vitamins, minerals, and various metabolites (Suman et al., 2015; De Vuyst et al., 2016; Onofre et al., 2017; Johansen et al., 2019; Tofalo et al., 2019). Thus, they contribute to a balanced diet in West Africa, where undernourishment affects 15% of the population (FAO, 2020).
In Burkina Faso, “Rabilé” is a popular traditional ferment that mainly contains yeasts. It is obtained at the end of sorghum beer production after a semi-controlled fermentation and corresponds to the dried lees (Konlani et al., 1996a, 1996b). Specific pathogenic and opportunistic yeasts can probably proliferate, and produce compounds that are harmful to the body (Steensels and Verstrepen, 2014; Johansen et al., 2019; Tofalo et al., 2019). On the other hand, this semi-controlled fermentation denotes a great diversity of wild yeasts from "Rabilé" with unexplored potential. Empirically, the incorporation of "Rabilé" in sauces and ready meals is similar to Single Cell Protein (SCP) as a food supplement or additive. We, therefore, believe that "Rabilé" is an alternative for the treatment and prevention of malnutrition.
For all these reasons, we hypothesize that “Rabilé” resulting from a process of semi-controlled fermentation constitutes a source of diversified wild yeasts potentially usable for food enrichment.
Yeast’s diversity analysis is mainly based on molecular genotyping. Esteve-Zarzoso et al. (1999) proposed a method of differentiating yeasts based on enzymatic restriction of the 5.8S-ITS region. This technique has been proven helpful in several diversity studies, particularly for yeasts in wine and beer (Granchi et al., 1999; Díaz et al., 2013; Corbett et al., 2019). However, to have a suitable level of discrimination for genetic analysis of populations, molecular techniques such as sequencing and microsatellites analysis are needed. This study aimed to identify the yeast microbiota in a traditional ferment, and to study their kinetic parameters.
MATERIALS AND METHODS
Sampling of “Rabilé” (traditional ferment)
Sampling was conducted in 13 sites representing the 13 administrative regions of Burkina Faso (Banfora, Bobo-dioulasso, Dédougou, Dori, Fada N’Gourma, Gaoua, Kaya, Kombissiri, Koudougou, Ouagadougou, Ouahigouya, Tenkodogo, Ziniaré). In each location, three local producers were randomly chosen from which one sample of “Rabilé” was collected (approximately 50 g). Samples collected were aseptically packaged in stomacher bag (Stomacher® 400 Classic Bags, Seward, UK), and transported under cold conditions (4°C).
Isolation of yeasts from samples of traditional ferments
To make serial decimal dilutions, 10 g of each sample were diluted in 90 mL of sterile physiological solution (0.9% NaCl). A volume of 100 µL of each dilution was inoculated on Sabouraud media with chloramphenicol. The set was incubated at 30°C for 48 to 72 h (Guimarães et al., 2006).
Well-individualized colonies were selected and purified by a series of three successive sub-culturing. A total of 30 isolates per site was selected, and stored at -20°C in Yeast Extract Peptone Dextrose Broth (YPD Broth) containing 20% glycerol for molecular and physiological characterization.
Molecular characterization of yeast isolates
PCR-RFLP
DNA was extracted from each yeast isolate using Cetyl Trimethylammonium Bromide (CTAB) buffer from a fresh culture (Kumar et al., 2014). The 5.8S-ITS region was amplified using the primer pair ITS1 (5’-TCCGTAGGTGAACCTGCGG-3') and ITS4 (5'TCCTCCGCTTATTGATATGC-3') as described by White et al. (1990). According to the manufacturer's instructions, PCR products were digested separately using three restriction enzymes HhaI, HaeIII, and HinfI (NEW ENGLAND Biolabs® Inc). The PCR products and their restriction fragments were separated on a 2.5% agarose gel. After electrophoresis, gel was photographed under ultra-violet light of the transilluminator (UVP®) coupled to a computer. All fragments were analyzed with UVP Doc-ItTM Ls Analysis software (Version 6.8.2) to determine their sizes. Isolates were then clustered according to their restriction profiles.
Sequencing of the 5.8S-ITS region
One strain was chosen from each restriction profile group for sequencing. The PCR products were purified (NEW ENGLAND Biolabs® Inc.) under a combined action of two hydrolytic enzymes (Exonuclease I and Shrimp Alkaline Phosphatase). The obtained products were re-amplified using the Genomelab-DTCS Quick Start Kit (Beckman Coulter, USA) following the cycle sequencing program of 30 cycles of denaturation (96°C, 20 s), annealing (50°C, 20 s), and extension (60°C, 4 min). Cycle sequencing products were purified by ethanol precipitation, and then separated using a CEQTM 8000 Genetic Analyzer Sequencer. Finally, the isolates' phylogenetic tree was constructed using the Geneious Prime software (Version 2020.0.3) from the sequences obtained using Tamura-Nei as Genetic Model Distance and Neighbor-Joining as Tree Build Method.
Single locus analysis
Three isolates of Saccharomyces cerevisiae were randomly selected per site to obtain 39 isolates for single-locus analysis. The ScAAT1 locus was amplified as described by Legras et al. (2005) using a pair of ScAAT1 primers represented by the ScAAT1 forward labeled (Cy5) and ScAAT1 reverse unlabeled. Using CEQTM 8000 Genetic Analyzer Sequencer and DNA standard size 400 bp, the amplified products were separated by capillary electrophoresis. GenAlex version 6.502 was used to determine diversity parameters.
Study of kinetic parameters
It consisted of monitoring the yeasts' growth in a non-renewed medium and determining their kinetic parameters by following the steps that follow.
Strains were first grown on YPD agar for 24 h. Then pure colonies obtained were inoculated into YPD broth incubated under continuous agitation overnight at 30°C to constitute a stock culture. Pre-culture was performed by inoculating 6 µl of stock culture into fresh YPD Broth incubated under continuous agitation overnight at 30°C. Cells were harvested from pre-culture by centrifuging at 3000 g for 2 min to constitute an inoculum of 1.38 × 107 cells/mL.
A volume of 10 µL (1.38×105 cells) of the inoculum (1.38×107 cells/mL) was transferred to the wells of a microplate (containing 290 µL of YPD Broth), except the three wells used as blank (control with no cells). The test was carried out in triplicate. The growth at 30°C was then monitored using a microplate reader spectrophotometer (Biotek ELx808, USA). The cellular biomass (Optical Density OD630) was recorded every 20 min for 24 h. A growth curve (OD as a function of time) was performed to determine the kinetic parameters: maximum specific growth rate (µmax) and generation time (tg). Analysis of variance was applied to assess significant kinetic differences.
RESULTS
390 yeast isolates were obtained from “Rabilé” samples collected from the 13 regions of Burkina Faso (Table 1).

Molecular characteristics of yeasts from “Rabilé”
Based on the size of the PCR products and the restriction patterns obtained, 390 yeast isolates were clustered into 13 genetic groups corresponding to 13 distinct restriction profiles (Table 1).
Similarity of sequences
Homology search results revealed 12 species among the 13 genetic groups with a similarity of 98.1 to 100% (Table 2).

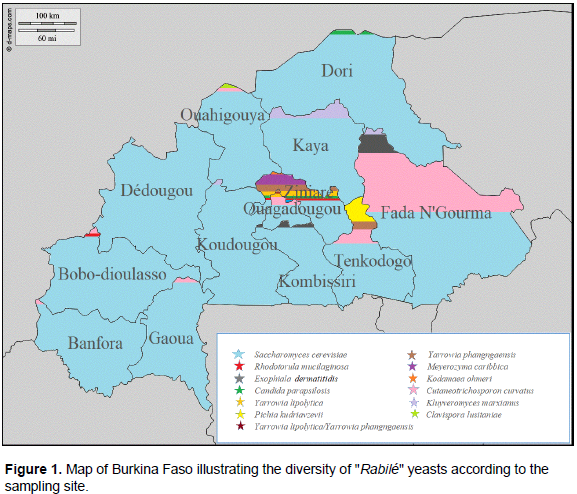
According to the sampling sites, we found seven species of yeast in samples from the Ziniaré site; five species in samples from Ouagadougou; four species in samples from Tenkodogo and Fada N’Gourma; three species in samples from Bobo-Dioulasso and Ouahigouya; two species in samples from Banfora, Dori, Gaoua, Kaya, Kombissiri, and Koudougou, and one species in samples from Dédougou (Figure 1). Figure 2 represents the Neighbor-Joining tree of these yeast species isolated from “Rabilé.”

A total of 38 Saccharomyces isolates were successfully analyzed using a single locus assay. We obtained 28 genotypes and 18 alleles with 0.902 as the Nei’s genetic diversity index for these isolates.
Kinetic parameters
The maximum specific growth rate is inversely proportional to the generation time, and is one of the most important parameters in modelling microbial growth (Barbera et al., 2019).
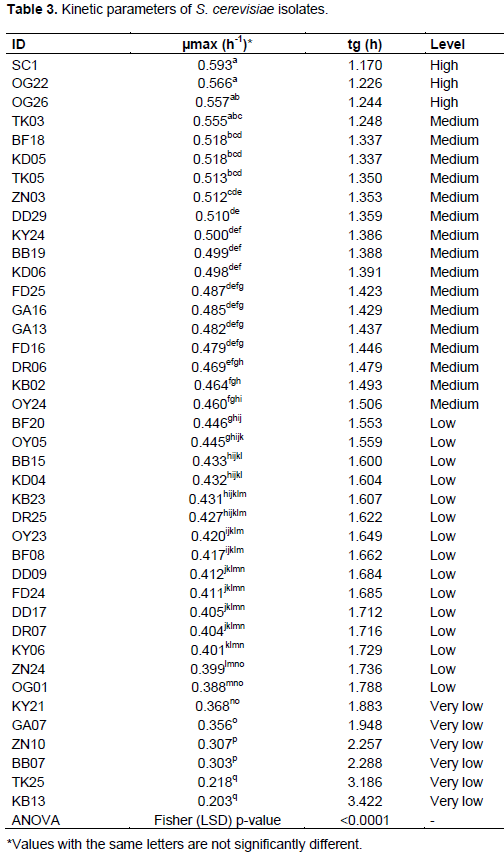
S. cerevisiae
The analysis of kinetic parameters showed that maximum specific growth rate (µmax) and generation time (tg) were significantly variable between isolates of S. cerevisiae. According to the classification of Oliveira et al. (2004), three of our isolates of S. cerevisiae (OG22, OG26, TK03) had a high µmax. Fifteen isolates reached a medium level. The isolate OG22 showed a high growth rate similar to that of the industrial strain used as a control (Table 3).
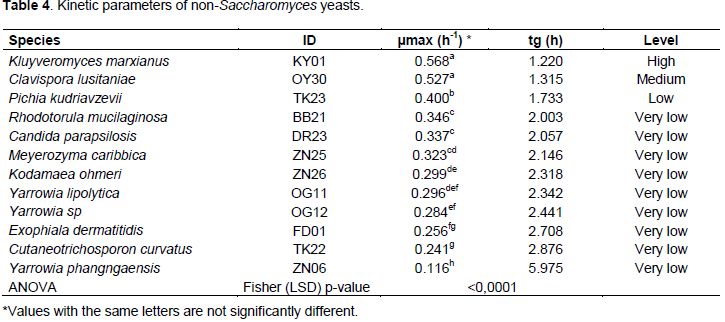
Non-saccharomyces species
Maximum specific growth rate and generation time were also significantly variable between species (Table 4). Kluyveromyces marxianus had a high maximum specific growth rate, while Yarrowia phangngaensis had the lowest.
DISCUSSION
This study highlights the “Rabilé” yeasts diversity from different production sites. It was hypothesized that “Rabilé” could contain a varied range of yeasts, as this starter results from uncontrolled alcoholic fermentation. It occurs in a non-sterile environment so that several microorganisms can intervene to enhance fermentation or as spoiling agents (Mogmenga et al., 2017). Nine yeast species identified in this study (S. cerevisiae, K. marxianus, Pichia kudriavzevii, Meyerozyma caribbica, Candida lusitaniae, Candida parapsilosis, Rhodotorula mucilaginosa, Yarrowia lipolytica, Kodameae ohmeri) are yeasts commonly encountered in fermented foods (N'guessan et al., 2011; Johansen et al., 2019). Some of these yeasts (Y. lipolytica, S. cerevisiae, and K. marxianus) belong to the list of microorganisms, and derived products approved by the Food and Drug Administration (FDA). It is reassuring that yeasts from traditional fermentations do not constitute a health risk for consumers in general (Steensels and Verstrepen, 2014).
Yeasts known to produce alcohol have been found in the ferment, including S. cerevisiae, K. marxianus, P. kudriavzevii, and M. caribbica (Djêgui et al., 2015; Tolieng et al., 2018; Johansen et al., 2019). These strains may be potential candidates for standardized starter production. Some species involved in organoleptic and nutritional quality improvement have been isolated from the traditional ferment "Rabilé". These yeasts (K. marxianus, R. mucilaginosa, and Y. lipolytica) would act with lactic acid bacteria to provide each production's sensory characteristics and health benefits (Sørensen et al., 2011; Calabretti et al., 2012; Johansen et al., 2019; de Souza Varize et al., 2019).
In contrast, other species reported as opportunistic pathogens (C. lusitaniae, C. parapsilosis, Exophiala) have been detected. These yeasts are associated with producing harmful substances, fungal infections, and even respiratory diseases (Pires et al., 2016). The presence of these opportunistic pathogenic yeasts in the
“Rabilé” could constitute a health concern, especially since the resulting sorghum beer “Dolo” is consumed directly without pasteurization.
There is no systematic link between a yeast's presence in a ferment, and its presence in the final product. N’guessan et al. (2011) pointed out that certain non-Saccharomyces yeast species appear sporadically, and may not appear in the final product. Nonetheless, non-Saccharomyces yeasts require safety tests beforehand (Steensels and Verstrepen 2014). Taxonomically, the data obtained indicate a predominance of hemi-ascomycetes. The oleaginous yeasts belonging to the group of hemi-ascomycetes appeared to be closer to the hemi-basidiomycetes present in “Rabilé” (Figure 2).
In contrast to previous studies, our findings prove that the yeast microbiota of “Rabilé” is more diverse with 11 additional species compared to two species (Konlani et al., 1996a, van der Aa Kühle et al., 2001). We are of the opinion that yeast microbiota from “Rabilé” could be more diverse in reality because the culture-dependent method used in this study for assessing yeast diversity relies on knowledge of the yeasts' characteristics, and the relative abundance of each species (Lv et al., 2013). Further research at the subspecies level, using single-locus analysis, showed a high diversity of S. cerevisiae from the same production site, and within the same sample. These isolates showed a high allelic and genotypical richness.
Thus, this study determines these wild yeasts' kinetic parameters for Saccharomyces and non-Saccharomyces species. There was a high kinetic variability depending on the strain or species of yeast used. Some of these yeasts had a high maximum specific growth rate close to an industrial strain used as a control in the study. These yeasts could constitute potential candidates for the improvement and the standardization of “Rabilé” production.
CONFLICT OF INTERESTS
The authors have not declared any conflict of interests.
ACKNOWLEDGEMENT
The authors are grateful to the training trainers’ program (Mali) and the West African Economic and Monetary Union for support.
REFERENCES
|
Barbera E, Grandi A, Borella L, Bertucco A, Sforza E (2019). Continuous cultivation as a method to assess the maximum specific growth rate of photosynthetic organisms. Frontiers in Bioengineering and Biotechnology 7:274. |
|
|
Bekatorou A, Psarianos C, Koutinas AA (2006). Production of Food Grade yeasts. Food Technology and Biotechnology 44(3):407-415. |
|
|
Calabretti A, La Cara F, Sorrentino A, Di Stasio M, Santomauro F, Rastrelli L, Gabrielli L, Limone F, Volpe MG (2012). Characterization of volatile fraction of typical Irpinian wines fermented with a new starter yeast. World Journal of Microbiology and Biotechnology 28(4):1433-1442. |
|
|
Carrau F, Gaggero C, Aguilar PS (2015). Yeast diversity and native vigor for flavor phenotypes. Trends in Biotechnology 33(3):148-154. |
|
|
Corbett KM, de Smidt O (2019). Culture-dependent diversity profiling of spoilage yeasts species by PCR-RFLP comparative analysis. Food Science and Technology International 25(8):671-679. |
|
|
de Souza Varize C, Christofoleti-Furlan RM, Muynarsk EDSM, de Melo Pereira GV, Lopes LD, Basso LC (2019). Biotechnological applications of nonconventional yeasts. In Yeasts in Biotechnology 46:107674. IntechOpen. |
|
|
De Vuyst L, Harth H, Van Kerrebroeck S, Leroy F (2016). Yeast diversity of sourdoughs and associated metabolic properties and functionalities. International Journal of Food Microbiology 239:26-34. |
|
|
Díaz C, Molina AM, Nähring J, Fischer R (2013). Characterization and dynamic behavior of wild yeast during spontaneous wine fermentation in steel tanks and amphorae. BioMed Research International, 2013. |
|
|
Djêgui KY, Kayodé APP, Tokpohozin ES, Gachomo EW, Kotchoni SO, Hounhouigan JD (2015). Phenotypic characters of yeasts isolated from kpete-kpete, a traditional starter of a Benin opaque sorghum beer. African Journal of Biotechnology 14(27):2227-2233. |
|
|
Esteve-Zarzoso B, Belloch C, Uruburu F, Querol A (1999). Identification of yeasts by RFLP analysis of the 5.8 S rRNA gene and the two ribosomal internal transcribed spacers. International Journal of Systematic and Evolutionary Microbiology 49(1):329-337. |
|
|
Fallah B, Zaini F, Ghazvini RD, Kachuei R, Kordbacheh P, Safara M, Mahmoudi S (2016). The antagonistic effects of Candida parapsilosis on the growth of Fusarium species and fumonisin production. Current Medical Mycology 2(1):1-6. |
|
|
FAO, IFAD, UNICEF, WFP, WHO (2020). The State of Food Security and Nutrition in the World 2020. Transforming food systems for affordable healthy diets. Rome, FAO. |
|
|
Feldmann H (2012). Yeast: Molecular and Cell Biology. 2nd edn. Wiley-Blackwell, Weinheim, Germany 464 p. |
|
|
Gómez-Pastor R, Pérez-Torrado R, Garre E, Matallana E (2011). Recent advances in yeast biomass production. In: D Matovic (ed.) Biomass - Detection, Production and Usage pp. 201-222. InTech, Rijeka, Croatia. |
|
|
Granchi L, Bosco M, Messini A, Vincenzini, M (1999). Rapid detection and quantification of yeast species during spontaneous wine fermentation by PCR-RFLP analysis of the rDNA ITS region. Journal of Applied Microbiology 87(6):949-956. |
|
|
Guimarães TM, Moriel DG, Machado IP, Picheth CM, Bonfim T (2006). Isolation and characterization of Saccharomyces cerevisiae strains of winery interest. Revista Brasileira de Ciências Farmacêuticas, 42(1):119-112. |
|
|
Johansen PG, Owusu-Kwarteng J, Parkouda C, Padonou SW, Jespersen L (2019). Occurrence and importance of yeasts in indigenous fermented food and beverages produced in sub-Saharan Africa. Frontiers in microbiology P 1789. |
|
|
Konlani S, Delgenès JP, Moletta R, Traore A, Doh A (1996a). Isolation and physiological characterization of yeasts involved in sorghum beer production. Food Biotechnology 10(1):29-40. |
|
|
Konlani S, Delgenès JP, Moletta R, Traore A, Doh A (1996b). Optimization of cell yield of Candida krusei SO1 and Saccharomyces sp. LK3G cultured in sorghum hydrolysate. Bioresource Technology 57(3):275-281. |
|
|
Kumar MS, Kaur G, Sandhu AK (2014). Genomic DNA Isolation from Fungi, Algae, Plant, Bacteria and Human Blood using CTAB. International Journal of Science and Research 3(9):617-618. |
|
|
Legras JL, Ruh O, Merdinoglu D, Karst F (2005). Selection of hypervariable microsatellite loci for the characterization of Saccharomyces cerevisiae strains. International Journal of Food Microbiology 102(1):73-83. |
|
|
Lv XC, Huang XL, Zhang W, Rao PF, Ni L (2013). Yeast diversity of traditional alcohol fermentation starters for Hong Qu glutinous rice wine brewing, revealed by culture-dependent and culture-independent methods. Food Control 34(1):183-190. |
|
|
Lyumugabe F, Gros J, Nzungize J, Bajyana E, Thonart P (2012). Characteristics of African traditional beers brewed with sorghum malt: a review. Biotechnologie, Agronomie, Société et Environnement 16(4):509-530. |
|
|
Mogmenga I, Somda KM, Keita I, Traore SA (2017). Evaluation of hygienic quality of ferment of local beer dolo used as condiment in Burkina Faso. African Journal of Biotechnology 16(26):1449-1456. |
|
|
N'guessan KF, Brou K, Jacques N, Casaregola S, Dje KM (2011). Identification of yeasts during alcoholic fermentation of tchapalo, a traditional sorghum beer from Côte d'Ivoire. Antonie Van Leeuwenhoek 99(4):855-864. |
|
|
Oliveira ES, Rosa CA, Morgano MA, Serra GE (2004). Fermentation characteristics as criteria for selection of cachaça yeast. World Journal of Microbiology and Biotechnology 20(1):19-24. |
|
|
Onofre SB, Bertoldo IC, Abatti D, Refosco D (2017). Chemical composition of the biomass of Saccharomyces cerevisiae-(Meyen ex EC Hansen, 1883) yeast obtained from the beer manufacturing process. International Journal of Advanced Engineering Research and Science 5(8):351-355. |
|
|
Pires RH, Brugnera MF, Zanoni MVB, Giannini MJS (2016). Effectiveness of photoelectrocatalysis treatment for the inactivation of Candida parapsilosis sensu stricto in planktonic cultures and biofilms. Applied Catalysis A: General 511:149-155. |
|
|
Somda M, Savadogo A, Barro N, Thonart P, Traore A (2011). Effect of minerals salts in fermentation process using mango residues as carbon source for bioethanol production. Asian Journal of Industrial Engineering 3(1):29-38. |
|
|
Sørensen LM, Gori K, Petersen MA, Jespersen L, Arneborg N (2011). Flavour compound production by Yarrowia lipolytica, Saccharomyces cerevisiae and Debaryomyces hansenii in a cheese-surface model. International Dairy Journal 21(12):970-978. |
|
|
Steensels J, Verstrepen KJ (2014). Taming wild yeast: potential of conventional and nonconventional yeasts in industrial fermentations. Annual Review of Microbiology 68:61-80. |
|
|
Suman G, Nupur M, Anuradha S, Pradeep B (2015). Single cell protein production: a review. International Journal of Current Microbiology and Applied Sciences 4(9):251-262. |
|
|
Tankoano A, Sawadogo-Lingani H, Savadogo A, Kabore D, Traore, Y (2017). Study of the process and microbiological quality of Gappal, a fermented food from Burkina Faso based on milk and millet dough. International Journal of Multidisciplinary and Current Research 5(1):104-110. |
|
|
Tofalo R, Fusco V, Böhnlein C, Kabisch J, Logrieco AF, Habermann D, Cho GS, Benomar N, Abriouel H, Schmidt-Heydt M, Neve H, Bockelmann W, Franz CMAP (2019). The life and times of yeasts in traditional food fermentations. Critical Reviews in Food Science and Nutrition 60(18):3103-3132. |
|
|
Tolieng V, Kunthiphun S, Savarajara A, Tanasupawat S (2018). Diversity of yeasts and their ethanol production at high temperature. Journal of Applied Pharmaceutical Science 8:136-142. |
|
|
van der Aa Kühle A, Jesperen L, Glover RL, Diawara B, Jakobsen M (2001). Identification and characterization of Saccharomyces cerevisiae strains isolated from West African sorghum beer. Yeast 18(11):1069-1079. |
|
|
White TJ, Bruns TD, Lee S, Taylor JW (1990). Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky J, White TJ (eds), PCR Protocols: a Guide to Methods and Applications. Academic Press, San Diego pp. 315-322. |
|
Copyright © 2024 Author(s) retain the copyright of this article.
This article is published under the terms of the Creative Commons Attribution License 4.0