ABSTRACT
Breast feeding has critical effects on the newborns and either mother’s health. Some of such health-improving effects of the mother’s milk is associated with the beneficial microbes, lactic acid bacteria (LAB), which are normally present in the mother’s milk. Thus, human milk-associated lactobacilli were isolated in this study and some of their probiotic properties was investigated. Afterwards, Lactobacillus strains were screened for low pH and bile acids tolerance. Molecular identification was carried out using 16SrDNA and polymerase chain reaction (PCR). Antibiotic resistance was evaluated with disk diffusion assay and the inhibitory effect of isolates on pathogenic bacteria was examined with well assay and zone inhibition. Isolation experiments resulted in 122 human milk- associated lactobacilli belonging to 12 species. The most dominant species was Lactobacillus casei followed by Lactobacillus fermentum, Lactobacillus plantarum and Lactobacillus gasser, respectively. Screening for probiotic properties showed that 19 isolates, belonging to, Lactobacillus, have interesting probiotic characteristics. The most prevalent antibiotic resistance was observed in case of vancomycin (63.15%) and no drug resistance was detected for chloramphenicol, penicillin, rifampin (0%). Three Lactobacillus strains, designated as L4, L14 and L16, were found as potential probiotic strains since they have indicated promising inhibitory effects against the studied pathogenic bacterial strains. Our results shed light on the considerable diversity of lactobacilli in human breast milk. Furthermore, the candidate probiotic strains detected in this research might be used as potential probiotic strains.
Key words: Breast milk, Lactobacillus, probiotics, inhibitory effect, polymerase chain reaction.
The name probiotic stemmed from the Greek “pro bios” meaning literally “for life” (Soccol et al., 2010). Breastfeeding, as the main source of infants’ nutrition, affects the development of the microbiota in the gastrointestinal tract (Martín et al., 2005). It is estimated that newborns consume around 0.8 L/day breast milk which contains 105-107 colony forming units (CFUs) of milk microorganisms (Olivares et al., 2006). Breast milk microbiota comprises an array of microbial groups including the genera of lactic acid bacteria (LAB) (Damaceno et al., 2017).
The most pervasive probiotic species are represented by the following genera: Lactobacillus, Streptococcus, and Bifidobacterium, but other microorganisms including enterococci and yeasts (e.g. Saccharomyces boulardi) have also been exploited as probiotics (Soccol et al., 2010). Among the organisms occurring in milk, some species including Lactobacillus salivarius, Lactobacillus fermentum, Lactobacillus gasseri, Bifidobacterium breve, Bifidobacterium adolescentis, and Bifidobacterium longum have shown potential to enhance mother and infant health (Ruiz et al., 2019). Lactobacilli are aerotolerant, Gram-positive, non-motile, non-spore-forming, catalase and oxidase negative bacilli that are mainly fermentative bacteria. They consume simple sugars and produce lactate as their main metabolic product. Many species of Lactobacillus have long been known as antagonists of pathogenic bacteria (Wadher et al., 2010). Lactobacillus species produce significant amounts of lactic acid which causes a detectable pH drop. The acidification influences the growth of various microorganisms in human gut. Furthermore, lactic acid accumulation affects the fungal growth. The other antimicrobial property of lactobacilli originates from the nisin, the small peptides produced by these small-genome bacteria (Lindgren and Dobrogosz, 1990). The protective effects of lactobacilli and other probiotics against various intestinal pathogens have been extensively studied over the past decades, leading to commercial usage of lactobacilli in probiotic market as an effective therapy for many gastrointestinal diseases for not only human, but also farm animals (Gronlund et al., 2000).
Gram-negative intestinal bacteria, especially genera Salmonella and Shigella as well as Escherichia coli, are known as the major food-borne infection and diarrhea in many developing countries including Iran (Fardsanei et al., 2018; Talebreza et al., 2016; Memariani et al., 2014). To make matters worse, the rise of antibiotic resistance is a growing concern worldwide. Probiotic may reduce the rate of development of drug resistant pathogenic strains secondary to widespread and injudicious antibiotic use (Soltan Dallal et al., 2016a).
Breast milk can be regarded as one of the main resources for potential probiotic candidates. Many studies showed the potential diversity of bacterial species of breast milk (Collado et al., 2009; Jost et al., 2013; Zimmermann and Curtis, 2020). LAB and Bifidobacterium make up to 85% of the entire bacterial population of the intestinal microflora (Soccol et al., 2010). Considering the positive effects of breast milk on the microflora of the gastrointestinal tract of newborns, lactobacilli were isolated from breast milk samples in this study. Furthermore, the capability of the Lactobacillus isolates to inhibit the gastrointestinal pathogens was investigated.
Sampling
One hundred breast milk samples, around 3 ml, were collected from volunteer breast feeder mothers who were between 22-36 years old. Volunteers had no antibiotic usage 2 weeks prior to sampling. Milk samples were collected in sterile conditions and promptly transported to the laboratory under refrigerated conditions. Briefly, before sample collection, wearing sterile gloves, the women cleaned the nipple and surrounding area with an alcohol swab to minimize the presence of skin bacteria. The first few drops of manually expressed milk were also discarded (Li et al., 2017).
Enrichment and isolation
Enrichments were performed in MRS medium with 10 times dilution. After 48 h of incubation at 37°C, aliquots were streaked on MRS agar plates which were also incubated at the above-mentioned conditions. The emerged colonies were further purified and catalase negative Gram-positive colonies were subjected to biochemical and morphological characterizations (Soltan Dallal et al., 2016b). L. plantarum strain PTCC 1058 was used as the positive control.
Carbohydrate fermentation
The MRS broth medium (pH = 8) with no glucose and neither meat extract, containing 0.004% chloramphenicol red was used for carbohydrate fermentation assays. Such experiments were performed using 13 sugars with 0.5% (w/v) concentration (Marroki et al., 2011). Microplates were inoculated with a dense suspension of the given isolates and coverage was performed by autoclave-sterilized paraffin to establish the anaerobic conditions. Incubation at 35°C was performed for 5 days and a color change from violet to red was assumed as carbohydrate fermentation positive (Davoodabadi et al., 2015).
Tolerance to acid and bile salts
To assess the tolerance of the isolates to acidic conditions, 1 ml of a 24 h culture of the isolate was inoculated into 9 ml of PBS buffer (pH = 2.5) and after 3 h of incubation at 37°C, the viability of the isolate was observed (Soltan Dallal et al., 2016c). Acid-resistant isolates were further screened for resistance against bile salts. Two test tubes, one containing 9 ml MRS broth with 0.3% (w/v) bile salt Oxgall and the other one containing 9 ml MRS broth without Oxgall (negative control) were defined for each acid-resistant isolate. Test tubes were inoculated with 90 µl of cultures of given isolates and incubated at 37°C. The optical density (OD600) of the test tubes was assessed at inoculation time and either after 8 h of incubation (Sharifi et al., 2017).
Antimicrobial properties
The antimicrobial properties of the acid-bile resistant lactobacilli against E. coli O157: H7 (ATCC 43894), Salmonella enteritidis (ATCC 14028), Shigella sonnei (ATCC 25931), Yersinia enterocolitica (ATCC 23715), methicillin-resistant Staphylococcus aureus (ATCC 43300) were determined using well diffusion method as described previously (Rammelsberg and Radler, 1990). The pathogenic bacteria were cultured first in Luria-Bertani (LB) broth (Merck Co. Darmstadt, Germany) for 24 h at 37°C and their concentration was adjusted to 107 CFUs/ml, then were cultured on the surface of nutrient agar medium (Merck Co. Darmstadt, Germany) employing well diffusion agar method. After growing on MRS broth liquid culture mediums in candle jar at 37°C for 20 h, centrifugation was performed at 13,000 rpm for 10 min. The culture supernatant was then collected. One hundred microliters of liquid on culture was added to each well of the nutrient agar plate using well diffusion agar method, and the plates were then incubated for 15 h at 37°C. After the incubation time, the diameter of the growth inhibition zone around the wells was measured. Isolates with no growth halo diameter or < 11 mm were defined as negative, 11-16 mm as a moderate inhibitor (+), 17-12 mm as strong inhibitor (++), and ≥ 23 mm as a very strong inhibitor (+++) (Jomehzadeh et al., 2020). L. rhamnosus GG was used as a positive control and MRS sterile broth was used as a negative control (Lashani et al., 2018).
DNA extraction and molecular methods
DNA extraction of Lactobacillus isolates for the molecular analysis was performed according to the method of Chandok et al. (2015) with some modifications. First, pure culture of Lactobacillus isolates was prepared on MRS agar medium and 2 to 3 colonies were dissolved in 50 μl of STE (Sodium Chloride-Tris-EDTA) solution. The suspension was placed in a water bath at 96°C for 10 min (Boiling method), then the suspension was centrifuged at 13,000 rpm for 3 min and the supernatant was used for PCR (Soltan Dallal et al., 2017a).
PCR for 16S rDNA gene was performed to confirm the identification of Lactobacillus isolates with probiotic potency. This reaction was performed with 12.5 μl of Master Mix, Forward and Reverse primers of 0.3 μl each, 2 μl of DNA, and final volume up to 25 μl by distilled water. For PCR reaction, primers 27F (5′-CTCGTTGCGGGACTTAA-3′) and 1522R (5′-GCAGCAGTAGGGAATCTTC-3′) were used (Soltan Dallal et al., 2017b). PCR was performed with initial denaturation at 94°C for 2 min, denaturation at 94°C for 30 s, binding at 55°C for 1 min, elongation at 72°C for 1 min, and final elongation at 72°C. It was performed for 10 min during 35 cycles. Amplified DNA fragments were observed as single bands with a size of 757 bp by agarose gel electrophoresis (1.5% w/v) stained with ethidium bromide (Figure 1).
Isolated bacterial species
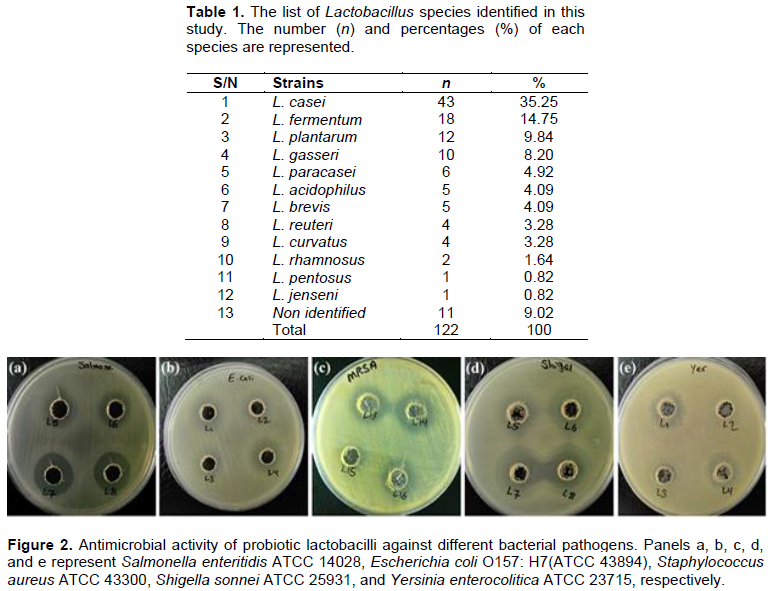
Out of 100 milk samples, 97 samples were positive for colony growth and the presence of bacillus and cocci lactic acid bacteria. A total of 122 Lactobacillus strains were isolated from breast milk samples. To identify Lactobacillus species, carbohydrate fermentation tests, growth test at 15 and 45°, arginine hydrolysis and glucose gas production were used. Among 122 isolates, 111 Lactobacillus species (90.99%) were identified by biochemical tests and 11 isolates (9.01%) were not identified during biochemical phenotypic tests. A total of 12 different species of Lactobacillus were identified in breast milk samples (Table 1). These species included: L. casei, L. fermentum, L. plantarum, L. gasseri, L. curvatus, L. paracasei, L. acidophilus, L. reuteri, L. brevis, L. pentosus, and L. rhamnosus. Among these species, the most dominant species was L. casei (n = 43; 35.25%).
Investigation of probiotic potency of isolates
Out of 122 Lactobacillus isolates studied, 22 isolates withstood acidic conditions with pH 2. In the next step, these 22 acid-resistant strains were examined for resistance to 0.3% oxgall bile salt. Of these isolates, 19 strains were finally resistant to both acidic and bile salt conditions and these strains were confirmed as strains with probiotic potential. A total of 19 Lactobacillus strains that showed resistance to acid and bile were L. casei (n = 5), L. brevis (n = 5), L. actobacillus (n = 2), L. fermentum (n = 2), L. paracasei (n = 2), L. rhamnosus (n = 2), and L. reuteri (n = 1).
Evaluation of antimicrobial effects of probiotic lactobacilli against gastrointestinal pathogens
Antimicrobial activity of 19 acid-resistant lactobacilli isolates against a number of gastrointestinal pathogens was examined (Figure 2). Antimicrobial activity of all 19 lactobacilli isolates was performed without temperature and enzymatic treatments. Out of 19 acid- and bile- resistant lactobacilli isolates, 10 strains (52.63%) were able to inhibit the growth of E. coli O157 H7, of which 6 strains had a strong inhibitory effect and 4 strains had a moderate inhibitory effect and 9 isolates did not have any inhibitory effect. The results showed that 12 isolates of probiotic lactobacilli (63.16%) were able to inhibit the growth of Salmonella, of which 9 isolates strongly or very strongly inhibited Salmonella. 3 isolates had moderate inhibitory effect and 7 isolates had no inhibitory effect. All 19 probiotic isolates (100%) were able to inhibit Shigella growth, of which 9 strains had strong effect, 6 strains had very strong effect and 4 strains had moderate inhibitory effect. 7 probiotic strains (36.84%) had inhibitory effect of S. aureus, of which 5 strains had a strong inhibitory effect and 2 strains had a moderate inhibitory effect. The other 12 strains of Lactobacillus had no inhibitory effect on the growth of S. aureus. 11 strains of probiotic Lactobacillus isolates (57.89%) had a strong inhibitory effect on the growth of Yersinia and 8 strains had no inhibitory effect on growth. Among 19 probiotic lactobacilli strains, 3 strains L4, L14 and L16 had strong inhibitory effect on all gastrointestinal pathogens tested.
The microflora of infants, especially lactic bacteria such as Lactobacillus and Bifidobacterium, plays a pivotal role in gastrointestinal tract in creating a proper balance of intestinal microorganisms, as well as beneficial properties on infant health and immune system promotion, in particular cellular immunity (Galdeano et al., 2019). The baby's gastrointestinal tract is sterile until birth, and microbial colonization begins gradually after this event. Microflora during infancy is under the influence of various factors including type of delivery (cesarean section or normal), environmental health, use of antibiotics by mother or baby, climatic and geographical conditions, type of feeding (formula or breast milk), and intestinal maturity (Navarro-Tapia et al., 2020). Lactobacilli are a heterogeneous group of lactic acid bacteria present in the intestinal flora of healthy people (Davoodabadi et al., 2015). These symbionts are part of the natural flora of the reproductive system of humans and animals. Breast milk is a continuous and useful food source for the development and creation of microorganisms in the baby's intestine, thereby identifying strains with probiotic properties from this source is of importance (Martín et al., 2005).
In this study, lactobacilli isolated from 100 samples of breast milk in which L. casei (35.2%) was the most abundant species. Bacteria growth was better during the isolation process under microaerophilic conditions. Therefore, it is recommended to use microaerophilic conditions for optimal isolation of these bacteria. Similar results have been reported by Brigidi et al. (2001).
In a study of conducted by Xanthopoulos et al. (1999), MRS was used to isolate lactobacilli at 37°C, which is in accordance with the method used in the present study. Yuki et al. (1999) also used lactitol-LBS-vancomycin agar (LLV) agar to isolate lactobacilli. The results of these studies show that in order to isolate lactobacilli, diverse but rich and slightly acidic culture media can be used (Xanthopoulos et al., 1999; Yuki et al., 1999).
In the present study, out of 122 strains, 11 strains (9%) could not be identified by phenotypic methods of carbohydrate fermentation. The difference between these bacteria in sugar fermentation and non-compliance with the standard tables could be due to mutations in one or more genes related to intermediate enzymes in fermentation of sugars that have mutated or deleted during evolution, change of environmental conditions and lack of need for the relevant gene (Hedberg et al., 2008).
Given that the identification of bacteria at the species level based on phenotypic characteristics is not accurate due to large changes in fermentation pattern, the study of ribosomal DNA gene sequences is a superior approach for phylogenetic comparison (Urbaniak et al., 2016). Soto et al. (2014) examined 160 samples of breast milk for the presence of Lactobacillus and Bifidobacterium and from 40.91% of the samples succeeded in isolating lactobacilli, the predominant species of which included L. salivarius, L. fermentum, and L. gasseri, respectively. Additionally, the isolation rate of lactobacilli and bifidobacteria was significantly lower in mothers treated with antibiotics than in healthy mothers. The above study clearly states the importance of the use of antibiotics and the effectiveness of breast milk flora from the digestive flora during lactation. In the present study, the isolation of lactobacilli from breast milk samples was 97%. In our study, in order to isolate maximum lactobacilli in breast milk flora, mothers who were treated with antibiotics for two weeks before sampling were excluded (Soto et al., 2014).
In a study which conducted on 7 mothers, Jost et al. (2013) isolated staphylococci, streptococci, bifidobacteria, and lactobacilli over three periods of 3-6, 14-9, and 25-30 days postpartum. Of the lactobacilli, only L. brevis and L. gasseri was isolated from the studied individuals, while in the present study, L. casei and L. fermentum were the most dominant species. These differences can be attributed to the differences in the flora of breast milk in different lactation periods and various geographical areas (Jost et al., 2013).
In a study by Gomez-Gallego , L. plantarum species was solely isolated from milk samples lactobacilli, while in the current study, 12 different species of Lactobacillus were isolated. This difference in isolation can be justified by regional differences because the breast milk microbiome is affected by specific factors including genetics, health status, maternal nutrition and geographical location (Taghizadeh et al., 2017; Jiang et al., 2016). Urbaniak et al. (2016) isolated Staphylococcus, Streptococcus, and Lactobacillus strain from 39 Canadian breast milk samples using 16SrDNA, of which Staphylococci formed the dominant species with 31% prevalence rate (Urbaniak et al., 2016).
In another survey, Kumar et al. (2016) evaluated 80 breast milk samples from 4 different geographic regions in Europe (Spain and Finland), Africa (South Africa) and Asia (China). The results of this study showed that breast milk in different regions shows a completely different pattern in terms of the presence of microbial flora (Kumar et al., 2016). Moreover, the results of a study on 133 breast milk samples from various regions of Taiwan and China demonstrated that different stages of lactation are effective in microbiome diversity and the composition of the microbial flora of breast milk can be different based on geographical regions (Li et al., 2017).
This study has some limitations, which should be addressed in future. For instance, in order to consider probiotics for human consumption, they should undergo in vitro and in vivo biosafety assessment. Biosafety assessment should take into account the nature of the organism being used, method of administration, level of exposure, health status of consumers and physiological functions they are called on to carried out (Sanders et al., 2010). The antibiotic resistance is also an important criterion for biosafety. The probiotic should not contain any transferable antibiotic resistance gene. Further investigation is also warranted for probiotic use in high risk human populations including severely immune-compromised individuals, neonates or hospitalized patients. Probiotics should not produce high amount of biogenic amines because these low molecular weight compounds can lead to various human ailments such as vomiting, hypertension, palpitations, and headache. In addition, the mucin degradation is an important criterion for biosafety assessment of probiotics. The probiotic should not degrade mucin (Kurkutia et al., 2019).
The present study on the study of breast milk microflora showed that there are various species of Lactobacillus in the milk of healthy mothers. The reason for this diversity of Lactobacillus species in breast milk is probably due to the type of nutrition and diet, lifestyle, age, climatic and geographical conditions, immune system status and many other factors.
Phenotypic identification methods based on biochemical properties alone are not sufficient to properly identify and differentiate all of the bacterial species; thereby more accurate techniques, in particular molecular methods, such as 16SrDNA gene sequencing are required for identification of various specific probiotic strains.
In our study, Lactobacillus species with good probiotic potential such as L. casei and L. fermentum were isolated, similar to other studies. Although various species of Lactobacillus were isolated from breast milk and even some species were dominant in terms of frequency, only few species showed probiotic properties. This study confirms that probiotic property is strain-dependent. Future studies should focus on the factors that may modulate the quantitative and qualitative composition of the breast milk microbiota.
The authors have not declared any conflict of interests.
The authors appreciate Microbiology Lab staff. This study was a part of a research project approved by the Food Microbiology Research Center, Tehran University of Medical Sciences, Tehran, Iran (Contract No. 29352).
REFERENCES
|
Brigidi P, Vitali B, Swennen E, Bazzocchi G, Matteuzzi D (2001). Effects of probiotic administration upon the composition and enzymatic activity of human fecal microbiota in patients with irritable bowel syndrome or functional diarrhea. Research in Microbiology 152(8):735-741.
Crossref
|
|
|
|
Chandok H, Shah P, Akare UR, Hindala M, Bhadoriya SS, Ravi GV, Sharma V, Bandaru S, Rathore P, Nayarisseri A (2015). Screening, isolation and identification of probiotic producing Lactobacillus acidophilus strains EMBS081 & EMBS082 by 16S rRNA gene sequencing. Interdisciplinary Sciences: Computational Life Sciences 7(3):242-248.
Crossref
|
|
|
|
|
Collado MC, Delgado S, Maldonado A, Rodríguez JM (2009). Assessment of the bacterial diversity of breast milk of healthy women by quantitative real?time PCR. Letters in applied microbiology 48(5):523-528.
Crossref
|
|
|
|
|
Damaceno QS, Souza JP, Nicoli JR, Paula RL, Assis GB, Figueiredo HC, Azevedo V, Martins FS (2017). Evaluation of potential probiotics isolated from human milk and colostrum. Probiotics and antimicrobial proteins 9(4):371-379.
Crossref
|
|
|
|
|
Davoodabadi A, Soltan Dallal MM, Lashani E, Tajabadi Ebrahimi M (2015). Antimicrobial activity of Lactobacillus spp. isolated from fecal flora of healthy breast-fed infants against diarrheagenic Escherichia coli. Jundishapur journal of microbiology 8(12):e27852.
Crossref
|
|
|
|
|
Fardsanei F, Soltan Dallal MM, Douraghi M, Memariani H, Bakhshi B, Zahraei Salehi T, Nikkhahi F (2018). Antimicrobial resistance, virulence genes and genetic relatedness of Salmonella enterica serotype Enteritidis isolates recovered from human gastroenteritis in Tehran, Iran. ournal of global antimicrobial resistance 12:220-226.
Crossref
|
|
|
|
|
Galdeano CM, Cazorla SI, Dumit JML, Vélez E, Perdigón G (2019). Beneficial effects of probiotic consumption on the immune system. Annals of Nutrition and Metabolism 74(2):115-124.
Crossref
|
|
|
|
|
Gomez-Gallego C, Garcia-Mantrana I, Salminen S, Carmen CM (2016). The human milk microbiome and factors influencing its composition and activity. Seminars in Fetal and Neonatal Medicine 21(6):400-405.
Crossref
|
|
|
|
|
Gronlund M, Arvilommi H, Kero P, Lehtonen O, Isolauri E (2000). Importance of intestinal colonisation in the maturation of humoral immunity in early infancy: a prospective follow up study of healthy infants aged 0-6 months. Archives of Disease in Childhood-Fetal and Neonatal Edition 83(3):F186-F192.
Crossref
|
|
|
|
|
Hedberg M, Hasslöf P, Sjöström I, Twetman S, Stecksén?Blicks C (2008). Sugar fermentation in probiotic bacteria-an in vitro study. Oral microbiology and immunology 23(6):482-485.
Crossref
|
|
|
|
|
Jiang M, Zhang F, Wan C, Xiong Y, Shah NP, Wei H, Tao X (2016). Evaluation of probiotic properties of Lactobacillus plantarum WLPL04 isolated from human breast milk. Journal of Dairy Science 99(3):1736-1746.
Crossref
|
|
|
|
|
Jomehzadeh N, Javaherizadeh H, Amin M, Saki M, Al-Ouqaili MTS, Hamidi H, Seyedmahmoudi M, Gorjian Z (2020). Isolation and identification of potential probiotic Lactobacillus species from feces of infants in southwest Iran. International Journal of Infectious Diseases 96:524-530.
Crossref
|
|
|
|
|
Jost T, Lacroix C, Braegger C, Chassard C (2013). Assessment of bacterial diversity in breast milk using culture-dependent and culture-independent approaches. British Journal of Nutrition 110(7):1253-1262.
Crossref
|
|
|
|
|
Kumar H, du Toit E, Kulkarni A, Aakko J, Linderborg KM, Zhang Y, Nicol MP, Isolauri E, Yang B, Collado MC, Salminen S (2016). Distinct patterns in human milk microbiota and fatty acid profiles across specific geographic locations. Frontiers in microbiology 7:1619.
Crossref
|
|
|
|
|
Kurkutia DK, Mistry N, Dwivedi M (2019). Probiotic properties and in vitro biosafety assessment of human Breast milk isolates. Journal of Pure and Applied Microbiology 13(2):1121-1134.
Crossref
|
|
|
|
|
Lashani E, Davoodabadi A, Soltan DMM (2018). Antimicrobial Effects of Lactobacillus Plantarum and Lactobacillus Paracasei isolated from honey against Staphylococcus aureus. Journal of Babol University of Medical Sciences 20(3):44-49.
|
|
|
|
|
Li SW, Watanabe K, Hsu CC, Chao SH, Yang ZH, Lin YJ, Chen CC, Cao YM, Huang HC, Chang CH, Tsai YC (2017). Bacterial composition and diversity in breast milk samples from mothers living in Taiwan and Mainland China. Frontiers in microbiology 8:965.
Crossref
|
|
|
|
|
Lindgren SE, Dobrogosz WJ (1990). Antagonistic activities of lactic acid bacteria in food and feed fermentations. FEMS microbiology reviews 7(1-2):149-163.
Crossref
|
|
|
|
|
Marroki A, Zúñiga M, Kihal M, Pérez- Martínez G (2011). Characterization of Lactobacillus from Algerian Goat'S Milk Based on Phenotypic, 16S rDNA Sequencing and their Technological Properties. Brazilian Journal of Microbiology 42(1):158-171.
Crossref
|
|
|
|
|
Martín R, Olivares M, Marín ML, Fernández L, Xaus J, Rodríguez JM (2005). Probiotic potential of 3 Lactobacilli strains isolated from breast milk. Journal of Human Lactation 21(1):8-17.
Crossref
|
|
|
|
|
Memariani M, Najar PS, Mostafavi SKS, Zahraei ST (2014). Detection of class 1 and 2 integrons among enteropathogenic Escherichia coli isolates Archives of Pediatric Infectious Diseases 2(4):e16372.
Crossref
|
|
|
|
|
Navarro-Tapia E, Sebastiani G, Sailer S, Toledano LA, Serra-Delgado M, García-Algar O, Andreu-Fernández V (2020). Probiotic supplementation during the perinatal and infant period: effects on gut dysbiosis and disease. Nutrients 12(8):2243.
Crossref
|
|
|
|
|
Olivares M, Díaz-Ropero MP, Gómez N, Lara-Villoslada F, Sierra S, Maldonado JA, Martín R, López-Huertas E, Rodríguez JM, Xaus J (2006). Oral administration of two probiotic strains, Lactobacillus gasseri CECT5714 and Lactobacillus coryniformis CECT5711, enhances the intestinal function of healthy adults. International journal of food microbiology 107(2):104-111.
Crossref
|
|
|
|
|
Rammelsberg M, Radler F (1990). Antibacterial polypeptides of Lactobacillus species. Journal of Applied Bacteriology 69(2):177-184.
Crossref
|
|
|
|
|
Ruiz L, García-Carral C, Rodriguez JM (2019). Unfolding the human milk microbiome landscape in the omics era. Frontiers in microbiology 10:1378.
Crossref
|
|
|
|
|
Sanders ME, Akkermans LMA, Haller D, Hammerman C, Heimbach J, Hörmannsperger G, Huys G, Levy DD, Lutgendorff F, Mack D, Phothirath P, Solano-Aguilar G, Vaughan E (2010). Safety assessment of probiotics for human use. Gut Microbes 1(3):164-185.
Crossref
|
|
|
|
|
Sharifi Yazdi MK, Davoodabadi A, Khesht Zarrin HR, Tajabadi Ebrahimi M, Soltan Dallal MM (2017). Characterisation and probiotic potential of lactic acid bacteria isolated from Iranian traditional yogurts. Italian Journal of Animal Science 16(2):185-188.
Crossref
|
|
|
|
|
Soccol CR, de Souza VLP, Spier MR, Medeiros ABP, Yamaguishi CT, Lindner JDD, Pandey A, Thomaz-Soccol V (2010). The potential of probiotics: a review. Food Technology and Biotechnology 48(4):413-434.
|
|
|
|
|
Soltan Dallal MM, Keshtvarz M, Zamani S, Shirazi M (2016a). Evaluation of anti-microbial activity of Lactobacillus acidophillus and Lactobacillus ruteri against entero-pathogens by in vitro and in vivo methods. Journal of Gorgan University of Medical Sciences 18(1):45-52.
|
|
|
|
|
Soltan Dallal MM, Hosseini M, Davoodabadi A, Rajabi Z, Zamaniahari S (2016b). Isolation and identification of lactic acid bacteria in traditional pickles and salted pickles from Tehran, Iran. Razi Journal of Medical Sciences 23(143):81-90.
|
|
|
|
|
Soltan Dallal MM, Khesht Zarrin HR, Tajabadi Ebrahimi M, Davoodabadi A, Hakimian MM, Sadrabadi AA, Sharifi Yazdi MK (2016c) Isolation and biochemical identification of potentially Probiotic lactic acid bacteria isolated from traditional yogurt in Yazd province. Tolooebehdasht 14(6):171-183.
|
|
|
|
|
Soltan Dallal MM, Zamaniahari S, Davoodabadi A, Hosseini M, Rajabi Z (2017a). Identification and characterization of probiotic lactic acid bacteria isolated from traditional persian pickled vegetables. GMS hygiene and infection control 12:1-7.
|
|
|
|
|
Soltan Dallal MM, Davoodabadi A, Abdi M, Hajiabdolbaghi M, Sharifi Yazdi MK, Douraghi M, Tabatabaei Bafghi SM (2017b). Inhibitory effect of Lactobacillus plantarum and Lb. fermentum isolated from the faeces of healthy infants against nonfermentative bacteria causing nosocomial infections. New Microbe new infections15:9-13.
Crossref
|
|
|
|
|
Soto A, Martín V, Jiménez E, Mader I, Rodríguez JM, Fernández L (2014). Lactobacilli and bifidobacteria in human breast milk: influence of antibiotherapy and other host and clinical factors. Journal of pediatric gastroenterology and nutrition 59(1):78-88.
Crossref
|
|
|
|
|
Taghizadeh M, Ghasemian SH, Poursina F (2017). Identification of Lactobacillus plantarum in Breast Milk. Research in Molecular Medicine (RMM) 5(4):50-60.
Crossref
|
|
|
|
|
Talebreza A, Memariani M, Memariani H, Shirazi MH, Shamsabad PE, Bakhtiari M (2016). Prevalence and antibiotic susceptibility of Shigella species isolated from pediatric patients in Tehran. Archives of Pediatric Infectious Diseases 4(1).
Crossref5
|
|
|
|
|
Urbaniak C, Angelini M, Gloor GB, Reid G (2016). Human milk microbiota profiles in relation to birthing method, gestation and infant gender. Microbiome 4(1):1-9.
Crossref
|
|
|
|
|
Wadher KJ, Mahore JG, Umekar MJ (2010). Probiotics: living medicines in health maintenance and disease prevention. International Journal of pharma and bio sciences 1(3):1-9.
|
|
|
|
|
Xanthopoulos V, Ztaliou I, Gaier W, Tzanetakis N, Litopoulou?Tzanetaki E (1999). Differentiation of Lactobacillus isolates from infant faeces by SDS?PAGE and rRNA?targeted oligonucleotide probes. Journal of applied microbiology 87(5):743-749.
Crossref
|
|
|
|
|
Yuki N, Watanabe K, Mike A, Tagami Y, Tanaka R, Ohwaki M, Morotomi M (1999). Survival of a probiotic, Lactobacillus casei strain Shirota, in the gastrointestinal tract: selective isolation from faeces and identification using monoclonal antibodies. International journal of food microbiology 48(1):51-57.
Crossref
|
|
|
|
|
Zimmermann P, Curtis N (2020). Breast milk microbiota: A review of the factors that influence composition. Journal of Infection 81(1):17-47.
Crossref
|
|